Hello, I’m trying to understand the detailed mechanism of spiking CNN.
(reference: NengoDL MNIST tutorial)
This is a part of the code:
neuron_type = nengo.LIF(amplitude=0.01)
...
# add the third convolutional layer
conv3 = nengo_dl.Layer(tf.keras.layers.Conv2D(filters=128, strides=2, kernel_size=3))(
conv2, shape_in=(12, 12, 64)
)
conv3_lif = nengo_dl.Layer(neuron_type)(conv3)
...
conv3_p = nengo.Probe(conv3, label="conv3_p")
conv3_p_filt = nengo.Probe(conv3, synapse=0.1, label="conv3_p_filt")
conv3_lif_p = nengo.Probe(conv3_lif, label="conv3_lif_p")
conv3_lif_p_filt = nengo.Probe(conv3_lif, synapse=0.1, label="conv3_lif_p_filt")
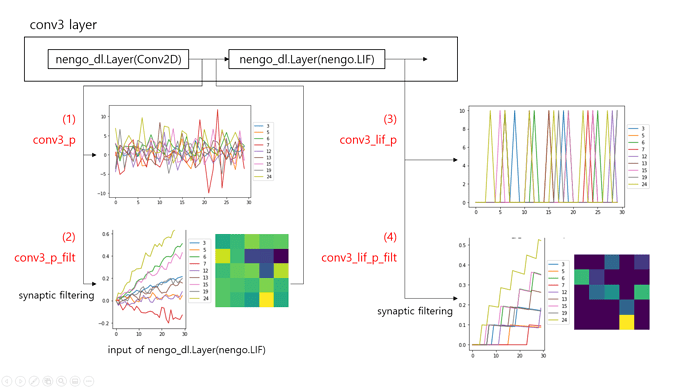
There are two nengo_dl.Layer objects in the third convolutional layer, and I probed data from each object for 30 timesteps with/without synaptic filtering. I got the following results.
[All situations are in the inference phase, not training phase]
-
From what I understand, LIF layer adds nonlinearity to the model and acts as a spiking version of activation function instead of non-spiking ReLU function. The filtered output from the Conv2D layer (2) becomes input to the LIF layer, inducing spikes of LIF neurons (3),(4)… But I have no idea how to interpret the output from Conv2D layer. Is Conv2D layer a group of spiking neurons? Where is the ‘synapse’ at which synaptic filtering takes place?
-
The final output of the second convolutional layer
conv2_lif_p_filtwould be similar to (4), then how does the thirdConv2Dlayer work? Does it apply convolution directly to theconv2_lif_p_filtdata with learned filters? -
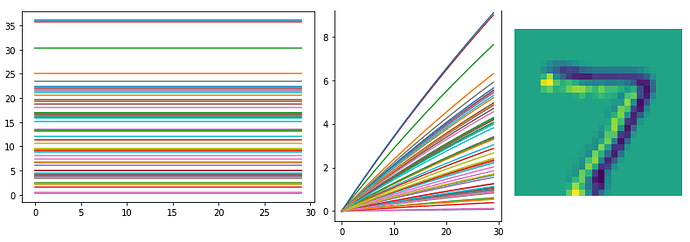
I also probed data from the first convolutional layer, and the output of Conv2D layer looks like the following. (left: without synaptic filtering (
conv1_p), right: with filtering (conv1_p_filt))
Is input image represented as an array of constant real values, similar to non-spiking CNN models?
Thank you in advance. Any references about details (such as synaptic filtering) are greatly welcomed.