Hello everyone,
I am trying to implement a one-layer fully connected SNN, in order to classify MNIST digits using STDP.
I was able to load the data using a Node and add the STDP rule to Nengo and I can see that the learning is happening as intended.
Now I want to add the Lateral inhibition (for the WTA) and neuron threshold adaptation (for the Homeostasis), in order to force the neurons to specify into different classes.
So, for example, using a simple two neurons ensemble and taking only two digits from the MNIST dataset.
For the Lateral inhibition i used the following approach to do it:
inhib_wegihts = np.full((n_neurons,n_neurons),-5)
inhib = nengo.Connection(
layer1.neurons,
layer1.neurons,
transform=inhib_wegihts)
but still I am getting an identical result of the both neurons learning just one class (even when i added the number of neurons)
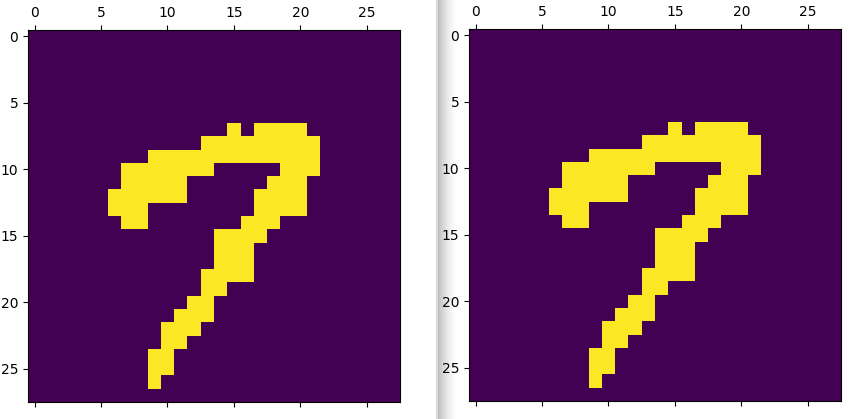
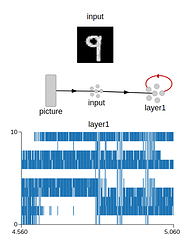
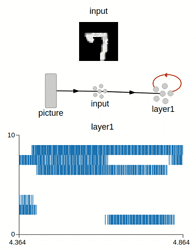
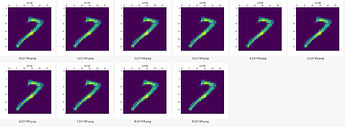
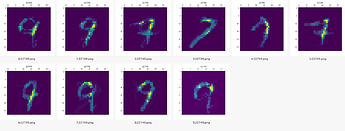
Preview of the weights after training (for 7 and 9):
For the threshold adaptation, I used the Adaptive LIF since (based on my understanding) it already contains an adaptive threshold based on the activity.
The full example (replaced STDP with BCM but still the same issue):
import nengo
import numpy as np
from numpy import random
from nengo_extras.data import load_mnist
#############################
# load the data
#############################
img_rows, img_cols = 28, 28
input_nbr = 1000
(image_train, label_train), (image_test, label_test) = load_mnist()
image_train = 2 * image_train - 1 # normalize to -1 to 1
image_test = 2 * image_test - 1 # normalize to -1 to 1
# select the 0s and 1s as the two classes from MNIST data
image_train_filtered = []
label_train_filtered = []
for i in range(0,input_nbr):
if label_train[i] == 7:
image_train_filtered.append(image_train[i])
label_train_filtered.append(label_train[i])
for i in range(0,input_nbr):
if label_train[i] == 9:
image_train_filtered.append(image_train[i])
label_train_filtered.append(label_train[i])
image_train_filtered = np.array(image_train_filtered)
label_train_filtered = np.array(label_train_filtered)
#############################
# Model construction
#############################
model = nengo.Network("My network")
presentation_time = 0.20 #0.35
#input layer
n_in = 784
n_neurons = 2
with model:
# input node
picture = nengo.Node(nengo.processes.PresentInput(image_train_filtered, presentation_time))
# input layer
input_layer = nengo.Ensemble(
784,
1,
label="input",
max_rates=nengo.dists.Uniform(22, 22)
)
# Connection between the node and input_layer
input_conn = nengo.Connection(picture,input_layer.neurons)
# layer
layer1 = nengo.Ensemble(
n_neurons,
1,
neuron_type=nengo.neurons.AdaptiveLIF(),
label="layer1",
)
# weights randomly initiated
layer1_weights = random.random((n_neurons, 784))
# Connection between the input and layer1
conn1 = nengo.Connection(
input_layer.neurons,
layer1.neurons,
transform=layer1_weights,
learning_rule_type=nengo.BCM(learning_rate=1e-4) # replaced later with the STDP rule
)
# Inhibition
inhib_wegihts = np.full((n_neurons,n_neurons),-10)
inhib = nengo.Connection(layer1.neurons, layer1.neurons, transform=inhib_wegihts)
with nengo.Simulator(model) as sim:
sim.run(presentation_time * label_train_filtered.shape[0])
So coming from another SNN event-driven simulator to Nengo, my questions are:
About my use-case:
1- Is there any issue with the way the inhibition is implemented in my example?
2- for the Homeostasis, is by using the AdaptiveLIF the right way to provide it?
About Nengo:
1- Nengo simulation uses dt, is it possible to implement event-driven simulations into Nengo? or is there an event-driven mode in Nengo maybe?
2- For the inhibition issue i had, i suspected maybe the default radius in Nengo is causing a problem since its by default between -1 and 1, while i used to work with a range from 0 to 1, it is possible to set it to that range? since i tried to modify it but there are always negative values involved in the interval.
Sorry for taking so long, i hope it is clear.
Have a nice day  .
.