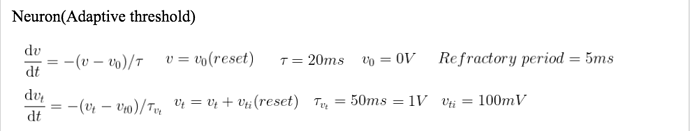Yes, that’s exactly what i used to do but in testing phase, you have basically two approaches : you set the labels at the end of testing phase based on either Max spikes or First spike and compute at the end the accuracy.
I had a doubt regarding implementing lateral inhibition.
inhib = nengo.Connection(
layer1.neurons,
layer1.neurons,
transform=inhib_wegihts)
What I instead wish to do, is to clamp the voltages of all other neurons to zero for 20ms. Any clue how can I do this?
You can emulate this by applying a synaptic filter on the lateral inhibition connection. Try a synapse of about 60ms to 80ms:
inhib = nengo.Connection(
layer1.neurons,
layer1.neurons,
transform=inhib_wegihts,
synapse=0.06)
Note that you may have to play with the inhibition weights to get it to respond quickly, or have multiple lateral inhibition connections with different synapses to span the 20ms window.
I would test what inhibition connection parameters would work for your requirements by creating a test network with just a feed-forward inhibition network, and then probing and plotting the output to see what effects changing the inhibition weights or synapse value does on the inhibited population.
Thanks
I did not get this. What do you mean by multiple lateral inhibition.
That would be of great help. Thanks
I shall try.
If you need to, you can actually create multiple lateral inhibition connections to get weird effects. For example, you could do:
with model:
inhib = nengo.Connection(
layer1.neurons,
layer1.neurons,
transform=inhib_weights)
inhib2 = nengo.Connection(
layer1.neurons,
layer1.neurons,
transform=inhib_weights,
synapse=0.01)
And you would get sort of 2 lateral inhibition events. You’ll have to experiment to see if this is the behaviour you’ll want in your code. Although, from my own experimentation, you’ll probably only need one lateral inhibition connection.
I’ve attached some test code here: test_inhib_synapse.py (2.0 KB)
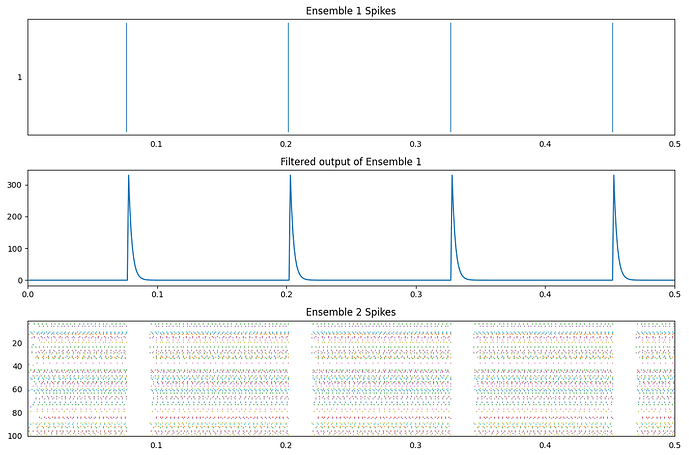
It looks like you should be able to get the 20ms inhibition delay with an inhibitory synapse of about 0.0025. The code has two ensembles, ens1 that inhibits ens2:
inhib_weight = 2
inhib_synapse = 0.0025
sim_runtime = 0.5
with nengo.Network() as model:
in_node = nengo.Node(1)
ens1 = nengo.Ensemble(
1,
1,
intercepts=nengo.dists.Choice([0]),
max_rates=nengo.dists.Choice([8]),
# We're using a low firing rate so that the spikes are far enough apart to
# demonstrate the 20ms inhibition of the ens2 spikes.
encoders=nengo.dists.Choice([[1]]),
)
ens2 = nengo.Ensemble(100, 1)
Note that this model is a pure feed-forward network just so that the only effects you see here are the direct inhibition effects and not a lateral inhibition. However, the same parameters should work with the lateral inhibition version (you’ll need to test it though).
Here’s an example output using the code:
And the code reports an average inhibition time of 20.86ms.
Yes this works. Thanks a lot.
I had one other doubt too.
Basically I am trying to move from brian2 to Nengo.
I am using neuron model :
dv/dt = -(v-v0)/tau: 1 (unless refractory) # membrane potential
dvt/dt = -(vt-vt0)*adaptation/tau_vt : 1 # adaptive threshold
tau : second # time constant for membrane potential
tau_vt : second # time constant for adaptive threshold
v = v0 # reset membrane potential
vt += vti # increment adaptive threshold
Synapse model :
Whenever pre synaptic spike occur, v+=w
I wish to reproduce the same behavior with refractory period of 2ms, tau of 30ms, tau_vt of 50ms, and vti of 0.1.
Can I use ensemble of such neurons (without using gain, bias, encoder, intercept and max firing rate)? Would it be possible to use such custom equations ?
I am using lateral inhibition, so modified transform to :
inhib_wegihts = (np.full((n_neurons, n_neurons), 1) - np.eye(n_neurons)) * 2
inhib = nengo.Connection(
layer1.neurons,
layer1.neurons,
synapse=0.0025,
transform=-inhib_wegihts*(n_neurons-1),
)
I hope that this is the right way to declare it?
Yes! You can make and use custom neuron classes in Nengo. Here’s an example on how do so: Adding new objects to Nengo — Nengo 4.0.1.dev0 docs
It is in general the right way to declare it, yes. I will note, however, that the notation I used in my example code:
transform=[[-inhib_weight]] * ens2.n_neurons
is python syntax to get the inhibition transform matrix into the right shape. When connecting to a .neurons object, the transformation matrix should be D \times n\\_neurons in shape, where D is the dimensionality of the source of the inhibitory signal. In my example code, the inhibitory signal is 1-dimensional (since it’s one neuron generating the inhibition signal), so the transformation matrix should be 1 \times \texttt{ens2.n_neurons} in size. In python, we can define such a matrix using [[value]] * ens2.n_neurons.
In the lateral inhibition code, because we are creating a feedback connection (connecting layer1 back on itself), the inhibition matrix should be n\\_neurons \times n\\_neurons in shape. This is what the np.full((n_neurons, n_neurons)) code does. However, since we want to do lateral inhibition and not self-inhibition, we subtract out the diagonal elements using - np.eye(n_neurons). Since the np.full function already creates a matrix of the right shape, you should only need transform=-inhib_wegihts instead of transform=-inhib_wegihts*(n_neurons-1).
Doing transform=-inhib_wegihts*(n_neurons-1) actually performs a scalar multiplication of the inhibition weight matrix with (n_neurons-1), so this makes your inhibition weight matrix dependent on the number of neurons you use in your layer1 population. If this is what you intended to do, you should note that the value used for transform will change the inhibition time per spike. Using my example code for example, if I modify my code such that inhib_weight is dependent on n_neurons, this is what I get for different values of n_neurons:
N_neurons: 1 , Average spike inhibition time: 0.02073684210526318
N_neurons: 2 , Average spike inhibition time: 0.022716666666666687
N_neurons: 5 , Average spike inhibition time: 0.024872340425531932
N_neurons: 10 , Average spike inhibition time: 0.026526315789473686
N_neurons: 20 , Average spike inhibition time: 0.027826086956521733
N_neurons: 30 , Average spike inhibition time: 0.029449999999999997
Thanks a lot for the detailed solution. Yes, I should not have multiplied with (n_neurons-1). Thanks for pointing out 
I don’t understand how to define a custom neuron with differential equations. Is there a way to do so?
This is the neuron model I am trying to implement.
The example I linked outlines the basic steps on how you would go about creating a custom neuron class. Essentially, you’ll want to sub-class the nengo.neurons.NeuronType class. In that class, the important function you’ll need to implement is the step() function, which contains all of the necessary neuron math.
As for neurons with differential equations, Nengo’s Izhikevich neuron implementation is an example of how you would accomplish this in Nengo. Here’s the link to the step function for reference: https://github.com/nengo/nengo/blob/master/nengo/neurons.py#L914
It was of great help. Thanks 
In the code you uploaded in lines 67 to 73 the following is stated
# Create a connection from pre to post
conn = nengo.Connection(
pre.neurons,
post.neurons,
transform=[[0]],
learning_rule_type=STDP(learning_rate=3e-6, bounds="none"),
)
What does transform=[[0]] do here exactly? What happens if I change this value? If I comment it out nengo states the following:
nengo.exceptions.ValidationError: Connection.learning_rule_type: Learning rule ‘STDP(bounds=‘none’, learning_rate=3e-06)’ cannot be applied to a connection that does not have weights (transform=None)
So it seems to have something to do with the weights. The docs state the following:
transform : (size_out, size_mid) array_like, optional
Linear transform mapping the pre output to the post input.
This transform is in terms of the sliced size; if either pre
or post is a slice, the transform must be shaped according to
the sliced dimensionality. Additionally, the function is applied
before the transform, so if a function is computed across the
connection, the transform must be of shape(size_out, size_mid).
But it doesn’t seem to really click for me.
To understand what is happening here, we need to look at the rest of the arguments in the nengo.Connection function call. Going line by line:
pre.neurons: This specifies that the connection will be made from the neurons of thepreensemble. Looking at the code, thepreensemble is created with 1 neuron in it.post.neurons: This specifies that the connection will be made to the neurons of thepostensemble. Looking at the code, thepreensemble is created with 1 neuron in it.learning_rule_type=STDP(...): This specifies that the connection will have anSTDPlearning rule used to modify the connection weights.
This error:
nengo.exceptions.ValidationError: Connection.learning_rule_type: Learning rule ‘STDP(bounds=‘none’, learning_rate=3e-06)’ cannot be applied to a connection that does not have weights (transform=None)
indicates that the STDP learning rule can only be applied to a connection that uses a connection weight matrix (i.e., the initial transform cannot be None). When creating a nengo.Connection that uses the STDP learning rule, we thus have to specify a value for the transform parameter.
But what value do we need to specify for transform? First, we need to figure out the shape of the transform. The value passed to the transform parameter should be a 2D array with a size of the destination by the size of the source. For the connection you are asking about, the transform should be a 2D array that is post.n_neurons by pre.n_neurons in shape. Since both ensembles have 1 neuron each, the transform should be [[x]] where x is some value. Note that using np.ones((post.n_neurons, pre.n_neurons)) * x is also valid.
And what should the value of x in the transform be? Well, that depends on you, the user. In my example script, I initialized the value of the connection weight between the two neurons to be 0 (to demonstrate that the STDP learning rule does actually modify it). However, depending on your use case, you could initialize it to some other value. As an example, I could configure the STDP example code to initialize it to a non-zero value, and then modify the spiking activity such that the STDP learning rule suppresses the connection weight value (i.e., by making the post spike before pre).
Putting both of these concepts together, we see why transform=[[0]] in my code.
Hello, I am seeking information on existing frameworks utilized for the conversion of Artificial Neural Networks (ANNs) to Spiking Neural Networks (SNNs). Several frameworks I have already come across include:
- SNN toolbox
- Sinabs
- SpikingJelly
- BindNET
- Spkeras
- NegoDL
- BrainCOG
- NEST
- ANNarcy
- Brain2
I would appreciate any additional suggestions beyond the ones listed above. Thank you