Hi @Eric,
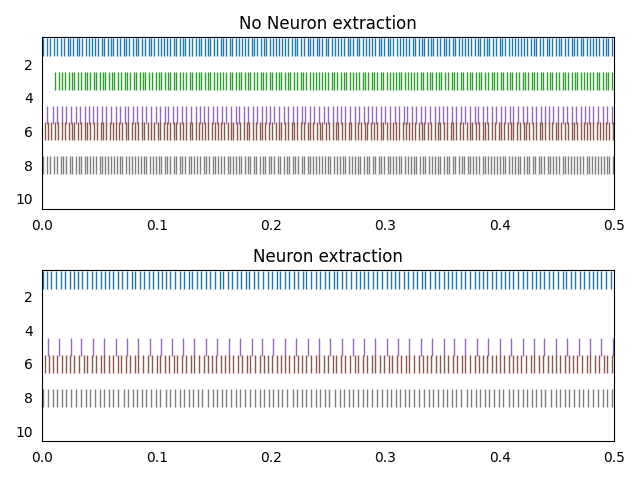
I printed the following parameters before and after I set weights to zero.
Before configuring weights = 0
- sim.data[ens2].bias
[ 11.33387593, -0.99492363, -30.79397568, -8.13427937, 3.07063806, 11.86735168, -1.94708691, 9.98872749, -3.41797826, -1.43711146]
- sim.data[ens2].max_rates
[356.6566124, 266.09879689, 326.87314282, 372.27112313, 242.11352655, 349.12747097, 288.7046451, 373.33176044, 202.66340248, 285.71903509]
- sim.data[ens2].gain
[14.05074736, 12.87879444, 50.1789458, 37.78255438, 6.82661282, 11.77680872, 16.11674386, 19.98729393, 10.74615897, 15.27721113]
- sim.data[ens2].intercepts
[-0.73546806, 0.15489987, 0.63361187, 0.24175918, -0.30331851, -0.9227756, 0.18285871, -0.44972208, 0.41112162, 0.15952594]
After configuring weights = 0
- sim.data[ens2].bias
[11.33387593, -0.99492363, -30.79397568, -8.13427937, 3.07063806, 11.86735168, -1.94708691, 9.98872749, -3.41797826, -1.43711146]
- sim.data[ens2].max_rates
[356.6566124, 266.09879689, 326.87314282, 372.27112313, 242.11352655, 349.12747097, 288.7046451, 373.33176044, 202.66340248, 285.71903509]
- sim.data[ens2].gain
[14.05074736, 12.87879444, 50.1789458, 37.78255438, 6.82661282, 11.77680872, 16.11674386, 19.98729393, 10.74615897, 15.27721113]
- sim.data[ens2].intercepts
[-0.73546806, 0.15489987, 0.63361187, 0.24175918, -0.30331851, -0.9227756, 0.18285871, -0.44972208, 0.41112162, 0.15952594]
As you can notice, there is no difference in Bias, Gain, Max Rates and Intercepts value which is understandable. The only difference between both of them is in the latter case, the weights = 0
Please note, I have set the seed value in nengo.Network and nengo.Ensemble. So in every execution of iris_v1.py, the bias, intercept, max rate and gain is the same.
Earlier you mentioned:
Just because the synapse weights between two ensembles are zero, does not mean that all the neurons in the second ensemble will be silent. Neurons in Nengo also have bias currents, which provide a “baseline” current in the absence of any input.
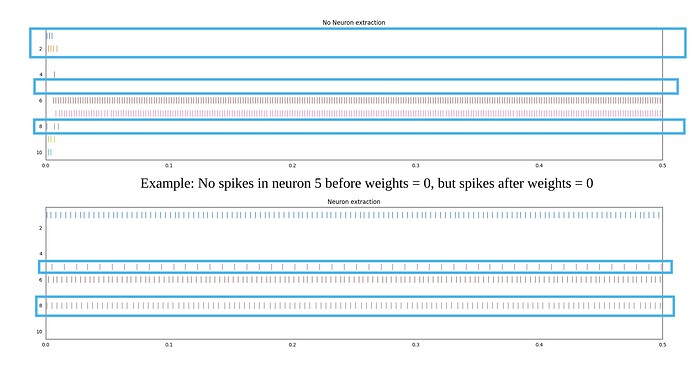
However, I see different neurons are activated in every execution and the spike rates are different before and after configuring weights = 0. (Please see the image uploaded(I have comments on the image). I have uploaded two images. Results of execution of iris_v1.py two times)
I understand why there are still spikes after weights = 0, but I do not understand
- why spike rates are different even though bias, gain, max rates, intercepts are same?
- Why the neuron which did not produce spikes before weights = 0 are now producing the spikes after weights = 0 even though bias, gain, max rates, intercepts are same?
In this link, it is mentioned that:
In Nengo, the behaviour of the neurons are dependent on the current input to the neuron.
I think by “current input”, it means the input that is fed to the code (in my case iris dataset). The input is the same in both the executions and sim.run. I am feeding the same dataset both the times. There are two sim.run (one before weights = 0 and one after weights = 0) in each execution.
Even if it means:
it should not change as gi, xi and bi are same before and after weights = 0 and in each executions.
I am just a little bid confused. What are the other factors that generate spikes? I believe it is not just the parameters in nengo.Ensemble. Am I correct?