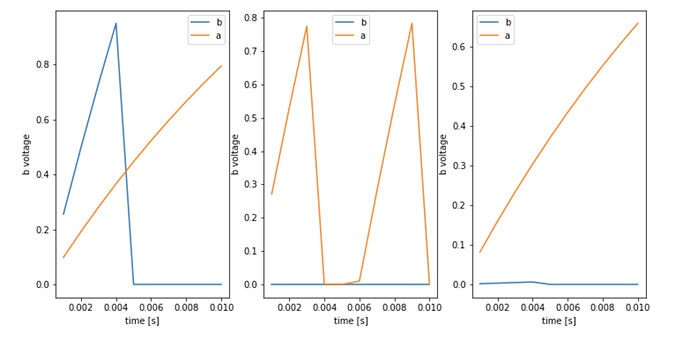
Hi,
I’m trying to understand the very basics of how connections work, but am having trouble finding exactly how a connection affects the voltage and current of the pre and post neurons. Is it in step_math of the neuron_type where the input from the previous layer is being used, or somewhere else? For example, if I make the following network, I am expecting that the input to b.neurons is the voltage array of a.neurons multiplied by the weights, which in this case, should be identical to the voltage of a.neurons. However, I don’t know where the inputs and outputs from probing the connection are coming from/what they mean:
weights = np.array([[1, 1, 1],
[1, 1, 1],
[1, 1, 1]])
model = nengo.Network()
with model:
a = nengo.Ensemble(3, dimensions=1, neuron_type=nengo.LIF())
b = nengo.Ensemble(3, dimensions=1, neuron_type=nengo.LIF())
conn = nengo.Connection(a.neurons, b.neurons, transform = weights)
spikes_probe = nengo.Probe(a.neurons, 'spikes')
voltage_probe = nengo.Probe(a.neurons, 'voltage')
b_spikes = nengo.Probe(b.neurons, 'spikes')
b_voltage = nengo.Probe(b.neurons, 'voltage')
conn_out = nengo.Probe(conn, 'output')
conn_in = nengo.Probe(conn, 'input')
with nengo.Simulator(model) as sim:
sim.run(.01)
plt.figure(figsize=(12, 6))
plt.subplot(131)
plt.plot(sim.trange(), sim.data[b_voltage][:,0], label = "b")
plt.plot(sim.trange(), sim.data[voltage_probe][:,0], label = 'a')
plt.xlabel('time [s]')
plt.ylabel('b voltage')
plt.legend()
plt.subplot(132)
plt.plot(sim.trange(), sim.data[b_voltage][:,1], label = "b")
plt.plot(sim.trange(), sim.data[voltage_probe][:,1], label = 'a')
plt.xlabel('time [s]')
plt.ylabel('b voltage')
plt.legend()
plt.subplot(133)
plt.plot(sim.trange(), sim.data[b_voltage][:,2], label = "b")
plt.plot(sim.trange(), sim.data[voltage_probe][:,2], label = 'a')
plt.xlabel('time [s]')
plt.ylabel('b voltage')
plt.legend()
sim.data[conn_out]
sim.data[conn_in]
I get the following graphs and values for conn_out and conn_in.