Hi all,
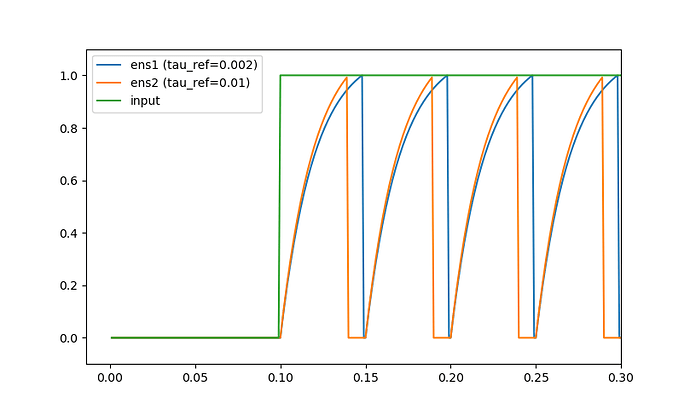
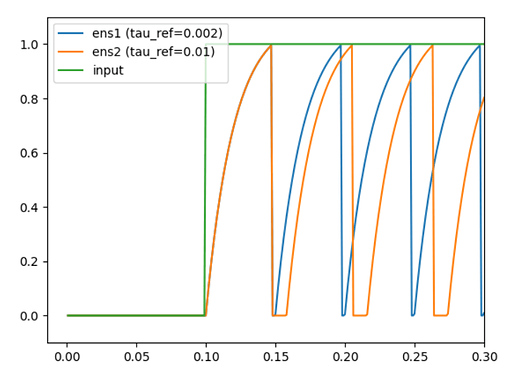
Comparing the voltages of single neurons, I am looking at how each parameter influences the spiking behaviour. From the documentation (both the parameter describtion as well as the step_math function) I expected that tau_ref would only affect the duration of the refractory period (i.e. the time that the voltage is kept at 0 after a spike.). However, for two neurons that are identical except for their tau_ref value, the neuron with a larger tau_ref reaches the spiking threshold faster (see figure). I do not see why this happens, as I expexted both neurons to reach the spiking threshold at the same time. Can someone explain to me why this is the case, that the potential increases quicker under the same input?
Code:
import numpy as np
import matplotlib.pyplot as plt
import nengo
model = nengo.Network(label="NEF summary")
def input_func(t):
if t < .1:
return 0
else:
return 1
with model:
input = nengo.Node(lambda t:input_func(t))
ens1 = nengo.Ensemble(
1,
dimensions=1,
intercepts=[0.99],
max_rates=[20],
encoders=[[1]],
label="ens1",
neuron_type=nengo.neurons.LIF(tau_ref=0.002),
seed=1
)
ens2 = nengo.Ensemble(
1,
dimensions=1,
intercepts=[0.99],
max_rates=[20],
encoders=[[1]],
neuron_type=nengo.neurons.LIF(tau_ref=0.01),
label="ens2",
seed=1
)
nengo.Connection(input, ens1, synapse=0)
nengo.Connection(input, ens2, synapse=0)
input_probe = nengo.Probe(input)
voltage_ens1 = nengo.Probe(ens1.neurons, "voltage")
voltage_ens2 = nengo.Probe(ens2.neurons, "voltage")
sim= nengo.Simulator(model)
sim.run(0.3)
t = sim.trange()
data_input = sim.data[input_probe]
data_ens1 = sim.data[voltage_ens1]
data_ens2 = sim.data[voltage_ens2]
# print("data_ens1:")
# print(data_ens1)
# print("data_ens2:")
# print(data_ens2)
plt.plot(t, data_ens1, label="ens1 (tau_ref=0.002)")
plt.plot(t, data_ens2, label="ens2 (tau_ref=0.01)")
plt.plot(t, data_input, label="input")
plt.xlim(right=t[-1])
plt.ylim(-0.1,1.1)
plt.legend()
plt.show()