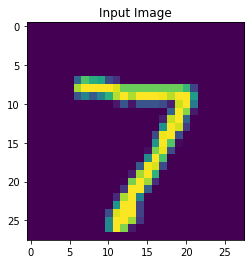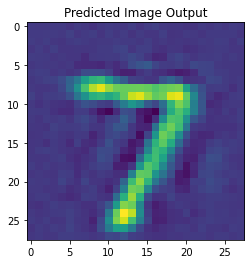Hi everyone,
Recently I’ve been playing around with a spiking Nengo autoencoder, which I was able to get working with the help of some of the great folks here (thanks for that!).
Here are some results from a model I trained to reconstruct neural spikes (trained on real neural data). The reconstructions are on the left and the original spikes are on the right.
I have two questions I’m thinking about now:
- In this example, each spike is a sample of 64 data points over a small window of time (so my input layer is 64 neurons). The inputs are fed into the network in parallel. However, I’d like to feed these data points (the spike waveform) in serial as a time sequence, and then reconstruct that time sequence to get the waveform. Would using the Legendre Memory Units be the best way to accomplish this, or is there some other way?
- The second thing I want to explore is the idea of “temporal compression” referenced in this thread: Dimensionality Reduction/Generative Modeling with Nengo
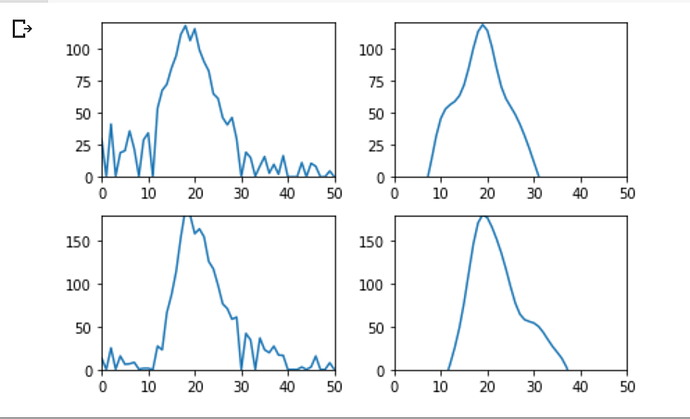
I’ve already seen some interesting results about how the length of the simulation affects reconstruction loss:
From left to right, the reconstructions are for 50 time steps, 100 time steps, and 150 time steps respectively.
Something I want to experiment with is for simulating different parts of the network for different amounts of time. For example, if I simulate the hidden layers for a shorter amount of time than the output layer, I could then learn a more temporally sparse representation of the input data, right?
Is there an easy way to do this with Nengo? If I did it by creating different networks, how do I connect between their ensembles so I can then maybe simulate one of them for a certain length of time steps and then another for a different number of time steps? Or would there be another way to do it?
Sorry if this is pretty long, just curious to hear people’s thoughts on this so I know what direction to go. Thanks!
 Let us know how things are progressing, this project is super cool!
Let us know how things are progressing, this project is super cool!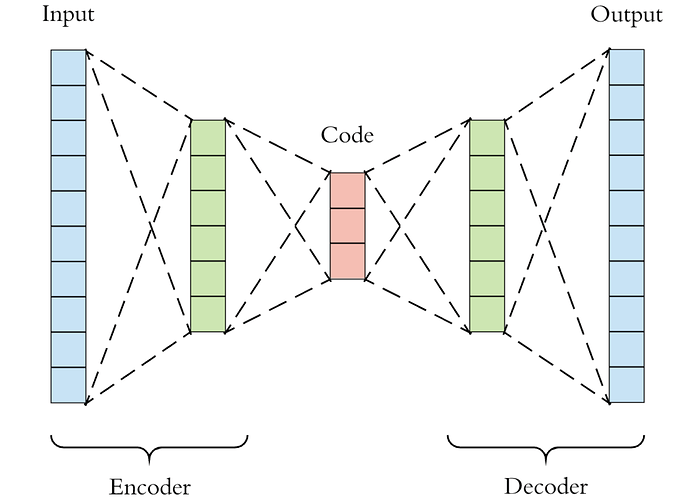
 )
)