Hi @Luciano,
You can “lesion” specific synapses in a Nengo network using a similar method to the neuron lesion code you linked in your post. First, it is important to note that in Nengo, a synapse is a filter applied to the connection between two ensembles. The synaptic filter is applied on the collective input to a neuron on the post population. This differs slightly from the biological definition of a synapse, which is between individual neurons. Thus, in the method I will outline below, to achieve the “lesioning” you desire, we modify the weights of the connection between the two ensembles, rather than modifying the synapse itself.
Similar to the “ablate neurons” function, a “lesion connection” function can be defined as follows:
def lesion_connection(sim, conn, lesion_idx):
connweights_sig = sim.signals[sim.model.sig[conn]["weights"]]
connweights_sig.setflags(write=True)
connweights_sig[lesion_idx, :] = 0
connweights_sig.setflags(write=False)
Note that the lesioning index code is dependent on how the nengo.Connection is created. If a connection between two ensembles is created like so (the default way):
conn = nengo.Connection(ensA, ensB)
The weights signal (i.e., sim.signals[sim.model.sig[conn]["weights"]]) has a shape that is (1, ensA.n_neurons). This means that it only contains the decoders of ensA. If you want to lesion the output of a neuron in ensA to every neuron in ensB it is connected to, no additional modification to the nengo.Connection is needed, and the lesion function can be used as is.
However, since you want to achieve the reverse (lesion all inputs to a specific neuron in ensB), we’ll need to change the code slightly. Namely, when we create the nengo.Connection, we specify the solvers with the weights=True flag to force the connection to be created with the full weight matrix. This weight matrix combines the decoders of ensA with the encoders of ensB. The full code for this is as such:
conn = nengo.Connection(ens1, ens2, solver=nengo.solvers.LstsqL2(weights=True))
With this change to the code, you can lesion the connection similar to how it was done with the neuron ablation code:
with nengo.Network() as model:
... # define your model
with nengo.Simulator(model) as sim:
lesion_connection(sim, conn, <lesion_index>)
sim.run(<runtime>)
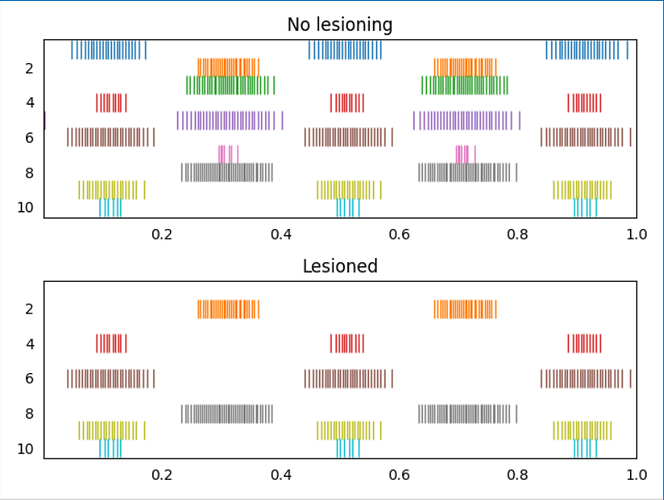
I’ve attached an example script (test_lesion_conn.py (1.5 KB)) that demonstrates this code. In the script, two ensembles are constructed, and here is the output plot of the script showing on non-lesioned run, and one lesioned run (the input connections to the 1st, 3rd, 5th, and 7th neurons in ens2 have been lesioned)
Some additional notes:
- The network is created with a seed so that multiple runs should be identical, which is why you see identical spike patterns for the non-lesioned connections.
- The lesioning is applied on a per-connection basis, so, you should be able to achieve the desired functionality of leaving other connections to “ens B” intact. You can test this out in the example code by adding an additional ensemble and connection to
ens2.