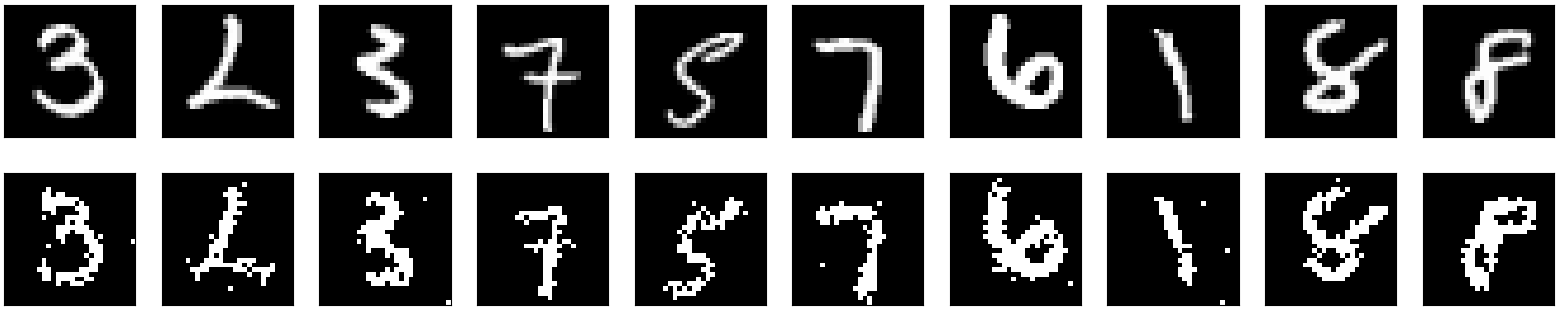@xchoo I have been going the the KerasSpiking examples and I tried applying it to a small model I have but I have not gotten any luck and I was wondering if you could take a look and tell me what is the problem.
I have a simple convolutional denoising auto encoder that uses the MNIST dataset. This is the network I have been using:
input = layers.Input(shape=(28, 28, 1))
# Encoder
x = layers.Conv2D(32, (3, 3), activation=tf.nn.relu, padding="same")(input)
x = layers.MaxPooling2D((2, 2), padding="same")(x)
x = layers.Conv2D(32, (3, 3), activation=tf.nn.relu, padding="same")(x)
x = layers.MaxPooling2D((2, 2), padding="same")(x)
# Decoder
x = layers.Conv2DTranspose(32, (3, 3), strides=2, activation=tf.nn.relu, padding="same")(x)
x = layers.Conv2DTranspose(32, (3, 3), strides=2, activation=tf.nn.relu, padding="same")(x)
out = layers.Conv2D(1, (3, 3), activation="sigmoid", padding="same")(x)
model = Model(inputs=input, outputs=out)
For the keras spiking model I changed the maxpooling layers to conv2d layers w/ stride 2 and changed it to follow the example in the documentation and this is what I came up with so far:
keras_spiking.default.dt = 0.01
LOWPASS = 0.1
filtered_model = tf.keras.Sequential(
[
# input
layers.Reshape((-1, 28, 28, 1), input_shape=(None, 28, 28)),
# conv1
layers.TimeDistributed(layers.Conv2D(32, (3, 3), padding='same', name='conv1')),
keras_spiking.SpikingActivation("relu", spiking_aware_training=True),
keras_spiking.Lowpass(LOWPASS, return_sequences=False),
# max pool
layers.TimeDistributed(layers.Conv2D(32, 2, 2, padding='valid', name='pool1')),
keras_spiking.SpikingActivation("relu", spiking_aware_training=True),
keras_spiking.Lowpass(LOWPASS, return_sequences=False),
# conv2
layers.TimeDistributed(layers.Conv2D(32, (3, 3), padding='same', name='conv2')),
keras_spiking.SpikingActivation("relu", spiking_aware_training=True),
keras_spiking.Lowpass(LOWPASS, return_sequences=False),
# max pool
layers.TimeDistributed(layers.MaxPooling2D((2, 2), padding='same', name='pool2')),
# deconv1
layers.TimeDistributed(layers.Conv2DTranspose(32, (3, 3), strides=2, padding='same', name='deconv1')),
keras_spiking.SpikingActivation("relu", spiking_aware_training=True),
keras_spiking.Lowpass(LOWPASS, return_sequences=False),
# deconv2
layers.TimeDistributed(layers.Conv2DTranspose(32, (3, 3), strides=2, padding='same', name='deconv1')),
keras_spiking.SpikingActivation("relu", spiking_aware_training=True),
keras_spiking.Lowpass(LOWPASS, return_sequences=False),
# output
layers.Conv2D(1, (3, 3), activation="sigmoid", padding="same")
]
)
However, this results in a dimension error in the first pooling layer:
ValueError: Input 0 of layer pool1 is incompatible with the layer: : expected min_ndim=4, found ndim=3. Full shape received: (None, 28, 32)
I tried flattening the input like in the keras spiking example but this lead to a dimension error in the first convolutional layer.
I’ve been looking for examples that involve using TimeDistributed with a Conv2D layer (like in this example here: TimeDistributed layer) but I tried do the same thing and I haven’t had any luck in successfully creating the model for training.
If you have the time I would greatly appreciate if you could look into why this model is not working with the keras spiking package. If you can’t find anything I can move onto modifying the source code like you said. I look forward to hearing back from you, thanks.