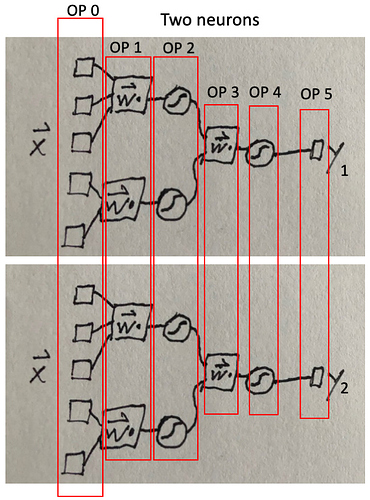
I’m trying to make networks with dendrite-like multi-layer perceptron features. I call these MLPs “dendrites” because they are tree graphs like many biological dendrites, and each neuron gets non-permutable inputs from heterogeneous “synapses,” unlike with weighted addition followed by a filter (i.e. nengo.Synapse).
It is perhaps related to BioSpaun from @jgosmann. The difference is that, being MLPs, they have nonlinearities but not conduction delays/states. It also might fall under a more traditional, weight-level neural modeling of the type described in @tbekolay’s post.
Sorry for the long and kind of vague post. I’m not sure if this is a Builder, Solver, or Nengo DL question. Any sort of partial advice would be helpful! I think it comes down to two parts.
1. Embedding stateless MLPs within a spiking network
Pseudocode:
def produce_one_neuron(*some_arguments):
with Network() as one_neuron:
dend_layer1 = Ensemble(4, 1, neuron_type=Sigmoid)
dend_layer2 = Ensemble(2, 1, neuron_type=Sigmoid)
soma = Ensemble(1, 1, neuron_type=LIF) # stateful neuron type
Connection(dend_layer1.neurons, dend_layer2.neurons, transform=L_4x2)
Connection(dend_layer2.neurons, soma.neurons, transform=L_2x1)
where L_4x2 means some 4x2 numpy array. See picture below.
The builder order of operations would look something like this pseudocode
one_neuron_steps = Simulator(produce_one_neuron()).step_order
one_neuron_steps == [TimeUpdate,
DotInc, SimProcess, # (the synapses)
DotSet, SimNeuron{Sigmoid}, # (dendrite layer 1)
DotSet, SimNeuron{Sigmoid}, # (dendrite layer 2)
DotInc, SimNeuron{SpikingSigmoid}] # (the soma/axon).
where DotSet is an imagined Y[...] = A.dot(X). I have already made the synapse Process - it is element-wise.
The point is that a spike arriving at a synapse at timestep i should be able to have an effect on the soma at timestep i.
It seems you could fold these “dendrites” into a fancy nengo.Neuron with some step_math composed of a bunch of Sigmoid functions; however, this would require a nengo.Neuron with a vector of inputs, instead of just scalar J. Is that possible?
Like the example above, another option is to make each layer of the MLP its own Ensemble; however, that ends up stacking timestep delays and internal states associated with the “DotInc” operators (hence why I made up the “DotSet” operators). Is there a way around that?
2. Getting the optimizer to recognize these patterns
Each dendritic subnet as well as each “synapse” Process are conceptually parallelizable – regardless of network topology – but the simulation optimizer does not recognize this if the neurons are constructed individually.
with Network() as multi_neuron:
for i in range(10):
produce_one_neuron()
The number of operations is theoretically constant in this case:
multi_neuron_steps = Simulator(multi_neuron).step_order
len(multi_neuron_steps) =approx= 10 * len(one_neuron_steps) # current behavior
len(multi_neuron_steps) == len(one_neuron_steps) # desired behavior
where basically all of the Dot steps should now be sparse BsrDot, just like what happens when you have a network of multiple Ensembles. Instead of step growth (bad), I’m looking for memory chunk growth (good).
I have tested both ways of initializing this type of network: neuron-by-neuron vs. layer-by-layer, and it made a big differece in speed - ridiculously big if you are using a GPU.
This seems like it will need a customization of either an operator or the optimizer. Both of these are pretty confusing to me. Any ideas/pseudocode on how to approach this?