Hello,
For a project I need to create LIF ensembles with various spiking threshold. As far as I know, it is not possible to choose this parameter in Nengo. Therefore, I changed the source code to provide the possibility of manually setting the threshold when creating a LIF:
`class LIF(LIFRate):
"""Spiking version of the leaky integrate-and-fire (LIF) neuron model.
Parameters
----------
tau_rc : float
Membrane RC time constant, in seconds. Affects how quickly the membrane
voltage decays to zero in the absence of input (larger = slower decay).
tau_ref : float
Absolute refractory period, in seconds. This is how long the
membrane voltage is held at zero after a spike.
min_voltage : float
Minimum value for the membrane voltage. If ``-np.inf``, the voltage
is never clipped.
amplitude : float
Scaling factor on the neuron output. Corresponds to the relative
amplitude of the output spikes of the neuron.
"""
probeable = ('spikes', 'voltage', 'refractory_time')
min_voltage = NumberParam('min_voltage', high=0)
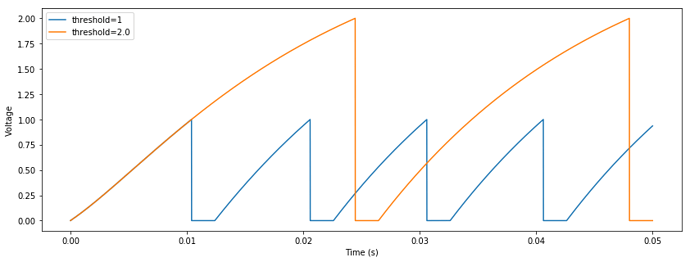
def __init__(self, tau_rc=0.02, tau_ref=0.002, min_voltage=0, amplitude=1, spiking_threshold=1):
super(LIF, self).__init__(
tau_rc=tau_rc, tau_ref=tau_ref, amplitude=amplitude)
self.min_voltage = min_voltage
self.spiking_threshold=spiking_threshold
def step_math(self, dt, J, spiked, voltage, refractory_time):
# reduce all refractory times by dt
refractory_time -= dt
# compute effective dt for each neuron, based on remaining time.
# note that refractory times that have completed midway into this
# timestep will be given a partial timestep, and moreover these will
# be subtracted to zero at the next timestep (or reset by a spike)
delta_t = (dt - refractory_time).clip(0, dt)
# update voltage using discretized lowpass filter
# since v(t) = v(0) + (J - v(0))*(1 - exp(-t/tau)) assuming
# J is constant over the interval [t, t + dt)
voltage -= (J - voltage) * np.expm1(-delta_t / self.tau_rc)
# determine which neurons spiked (set them to 1/dt, else 0)
spiked_mask = voltage > self.spiking_threshold
spiked[:] = spiked_mask * (self.amplitude / dt)
# set v(0) = 1 and solve for t to compute the spike time
t_spike = dt + self.tau_rc * np.log1p(
-(voltage[spiked_mask] - self.spiking_threshold) / (J[spiked_mask] - self.spiking_threshold))
# set spiked voltages to zero, refractory times to tau_ref, and
# rectify negative voltages to a floor of min_voltage
voltage[voltage < self.min_voltage] = self.min_voltage
voltage[spiked_mask] = 0
refractory_time[spiked_mask] = self.tau_ref + t_spike`
I basically replaced every 1 by self.spiking_threshold in step_math, although I’m not sure about what exactly is computed.
Although it successfully sets the threshold, this solution seems to affect the way the ensemble represents values. With the simple code below, the decoded value is inversely scaled by the given spiking_threshold:
`with model:
stim = nengo.Node([0])
a = nengo.Ensemble(n_neurons=50, dimensions=1,
neuron_type=nengo.neurons.LIF(spiking_threshold=2.))
nengo.Connection(stim, a)`
Is there a clean way to choose the spiking threshold? Can you give me some hints to implement it otherwise?