Understanding your model
I’m going to try and explain what you said back to you to make sure I understand.
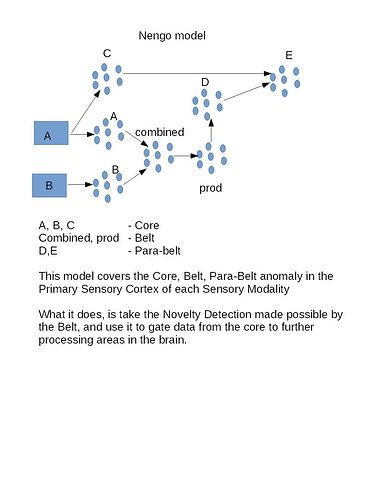
There are three cortical sections that seem important. They are the Core, Lateral Belt and Parabelt. You want to model the Lateral Belt as a Novelty Detector of data coming from the Core. You’ve defined Novelty as being the similarity between the Core and the Parabelt, which you’ve modeled as the product between two signals.
What behaviour do you want to initiate once you have this similarity measure? Alternatively, is it the ability to calculate the similarity that you are currently stuck on?
Understanding your code
Looking at your code, there’s a few problems and a few quality of life improvements that could be made. I’ve posted your code below for ease of reference:
import numpy as np
import matplotlib.pyplot as plt
import nengo
from nengo.processes import Piecewise
model = nengo.Network()
with model:
stim_a = nengo.Node(7.5)
stim_b = nengo.Node(Piecewise({0: .5, .1: 7.5, .2: .2}))
# set up the input neurons
a = nengo.Ensemble(n_neurons=100, dimensions=1, radius=1)
b = nengo.Ensemble(n_neurons=100, dimensions=1, radius=1)
c = nengo.Ensemble(n_neurons=100, dimensions=1, radius=1)
d = nengo.Ensemble(n_neurons=100, dimensions=1, radius=1)
e = nengo.Ensemble(n_neurons=100, dimensions=1, radius=1)
# set up the alternate path
combined = nengo.Ensemble(n_neurons=200, dimensions=2, radius=1.5)
prod = nengo.Ensemble(n_neurons=100, dimensions=1, radius=1)
combined.encoders = nengo.dists.Choice([[1, 1], [-1, 1], [1, -1], [-1, -1]])
nengo.Connection(stim_a, a)
nengo.Connection(stim_b, b)
nengo.Connection(stim_a, c)
# connect the alternate path
nengo.Connection(a, combined[0])
nengo.Connection(b, combined[1])
# connect the input neurons to the 2D neuron
# nengo.Connection(c, d[0])
nengo.Connection(d, e, transform=2.5)
# connect the Input Node to the alternate pathway
def product(x):
return x[0] * x[1]
# define the product function
nengo.Connection(combined, prod, transform=-2.5, function=product)
# connect up the product transform
threshold = -0.8
def thresh(x):
return x[0] - threshold
# define cross dimensional addition
# nengo.Connection(prod, d[1])
nengo.Connection(prod, d, transform=1.5, function=thresh)
nengo.Connection(c, e, transform=-.14) # , function=additn)
# connect up the addition transform
product_probe = nengo.Probe(prod, synapse=0.01)
d_probe = nengo.Probe(d, synapse=0.01)
e_probe = nengo.Probe(e, synapse=0.01)
# set up the probes to gather data
with nengo.Simulator(model) as sim:
sim.run(0.5)
# set up the simulator run
np.savetxt("prod_probe.txt", sim.data[product_probe])
np.savetxt("d_probe.txt", sim.data[d_probe])
np.savetxt("e_probe.txt", sim.data[e_probe])
# save the probes as txt type data files
plt.figure()
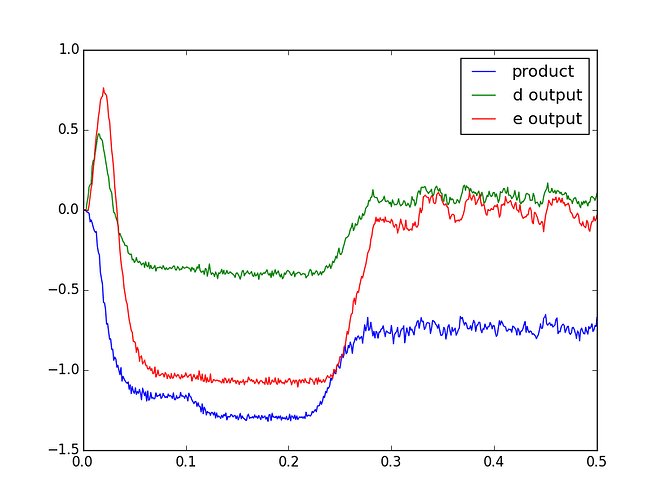
plt.plot(sim.trange(), sim.data[product_probe], label='product')
plt.plot(sim.trange(), sim.data[d_probe], label='d output')
plt.plot(sim.trange(), sim.data[e_probe], label='e output')
plt.legend()
plt.show()
What exactly are the inputs and outputs that you’re expecting from this network and how are they differing from what you’re currently getting?
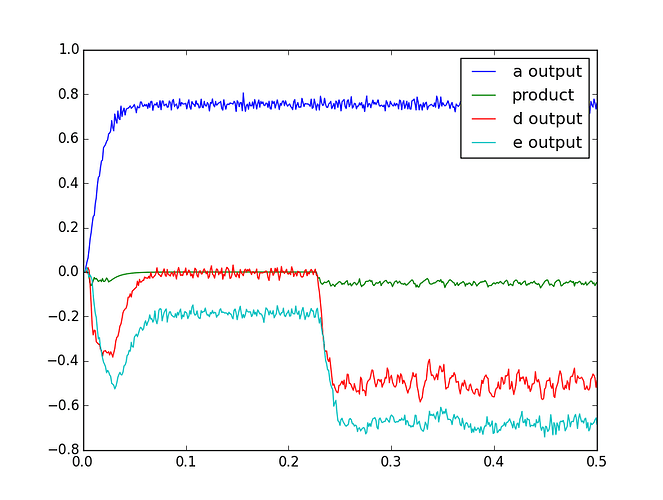
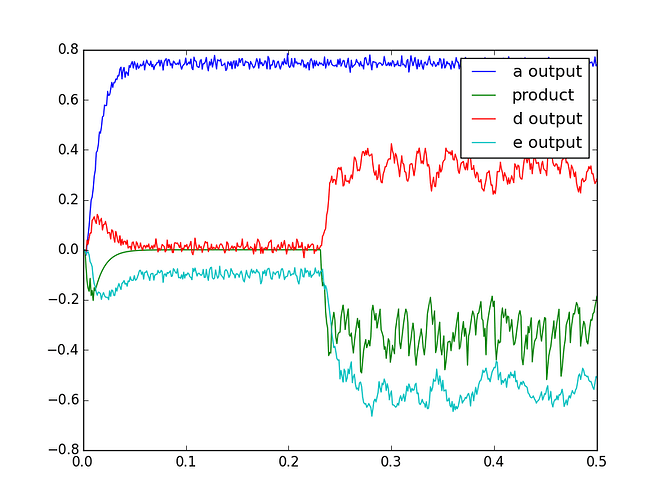
Even without totally understanding what’s happening in the code, there’s a few quirks worth noting. In the code, you’re using a custom product ensemble, but may I suggest using nengo.networks.Product instead? Additionally, you seem to want a to have a similarity threshold for some decision to be made. Your current implementation won’t work as you expect and I recommend instead using the configuration shown in this example. Finally, your inputs 7.5 and 7.5 greatly exceed the radius of the ensemble they are feeding into and might be the source of some of your confusion. These inputs are going to saturate the neural ensembles and are instead going to represent something around 1.2 instead of 7.5.
Bonus tips
One last thing. I’m a bit surprised you’re doing manual plotting. Have you tried using the Nengo GUI for exploring your model?