Hi ,
I have just installed nengo_ocl , and I was trying to run a a network of custom neurons I made with name (AFRN).
Then I got some error : " ‘Simulator’ object has no attribute ‘plan_SimAFRN’ ".
Am I doing something wrong? How to fix this?
Note that I added the neuron as a module in the same file of the network code only but didn’t add to the library.
Thanks in advance
Hi Omar,
Unfortunately nengo_ocl doesn’t know how to deal with custom neuron types. Currently, we manually specify an OpenCL kernel for all the parts of a Nengo simulation. For example, this pull request shows what we needed to add in order to get the Sigmoid and RectifiedLinear neuron classes working in nengo_ocl. You could do something similar for your neuron type if you know how to write OpenCL kernels.
There is a system for automatically translating Python code to OpenCL (see ast_conversion.py), but it’s limited in what it can convert, and we currently only use it for nodes. It could be possible to use it for custom neuron types as well; @Eric would be the person to talk to about that!
Thanks for your reply,
I would be grateful if it is possible to provde an example of how to use ast_conversion.py as I have no idea how to specify an OpenCL kernel.
Can you post your neuron code?
ast_conversion.py might work; it’s going to depend a lot on what exactly is happening in your neuron function.
But I think the easier route would be to just write the code yourself. You don’t actually have to write the whole kernel. You just have to write C code that does the computation for one neuron, and then I have code to vectorize it for many neurons. (See nengo_ocl.clra_nonlinearities.plan_rectified_linear for a simple example, and nengo_ocl.clra_nonlinearities.plan_lif for a more complicated one.)
Hi @Eric ,
Thanks for your concern
Here is my neuron code
import numpy as np import nengo from nengo.builder import Builder from nengo.builder import Operator from nengo.params import NumberParam
class AFRN(nengo.neurons.NeuronType): # Neuron types must subclass `nengo.Neurons` """A rectified linear neuron model.""" omega = NumberParam('omega', low=-0.0, low_open=False) g = NumberParam('g', low=0, low_open=True) thre = NumberParam('thre', low=0, low_open=True)
def __init__ (self,omega=1.0,g=0.9,thre=0.4): super(AFRN, self).__init__() self.omega = omega self.g = g self.thre = thre @property def _argreprs(self): args = [] if self.omega != 0.0: args.append("omega=%s" % self.omega) if self.g != 0.9: args.append("g=%s" % self.g) if self.thre != 0.4: args.append("thre=%s" % self.thre) return args def gain_bias(self, max_rates, intercepts): """Return gain and bias given maximum firing rate and x-intercept.""" gain = np.ones_like(max_rates) bias = np.zeros_like(max_rates) self.gain = gain self.bias = bias return gain, bias def rates(self, x, gain, bias): """Always use LIFRate to determine rates.""" J = x out = np.zeros_like(J) AFRN.step_math(self, dt=0.2, J=J, output=out) return out
def step_math(self, dt, J, output): si_out=[] omega = self.omega g = self.g thre = self.thre x = J if x.shape[0] == len(self.gain): for i in range(0,len(x)): si = np.tanh(g*(x[i]+omega*output[i])) if si > thre: si_out.extend([si]) elif si < thre: si_out.extend([0.]) output[...] = si_out else: output[...] = x
class SimAFRN(Operator):
def __init__(self, neurons, J, output, states=None, tag=None): super(SimAFRN, self).__init__(tag=tag) self.neurons = neurons self.J = J self.output = output self.states = [] if states is None else states
self.sets = [output] self.incs = [] self.reads = [J] self.updates = []
def _descstr(self): return '%s, %s, %s' % (self.neurons, self.J, self.output)
def make_step(self, signals, dt, rng): J = signals[self.J] output = signals[self.output]
def step_simafrn(): self.neurons.step_math(dt, J, output) return step_simafrn
@Builder.register(AFRN) def build_afrn(model, neuron_type, neurons): model.operators.append(SimAFRN( output=model.sig[neurons]['out'], J=model.sig[neurons]['in'],neurons=neuron_type))
The neuron function part is :
for i in range(0,len(x)):
si = np.tanh(g*(x[i]+omega*output[i]))
if si > thre:
si_out.extend([si])
elif si < thre:
si_out.extend([0.])
Which is built upon G.Edelman’s work:
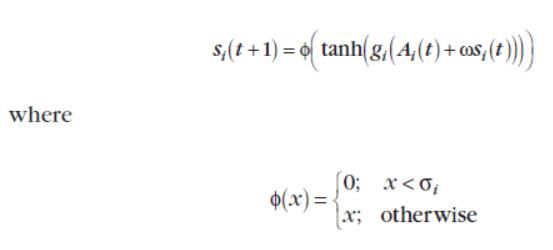
I tried to put the function without the part:
else:
output[…] = x
but I got dimensional error, that why I added this part for the first run (I hope it does not affect the accuracy of solution)
I hope my explanation was clear enough…
Hi,
I tried to do as explained in the example. I modified clra_nonlinearities.py and simulator.py. But when I try to run still gives the same error as before:
AttributeError: ‘Simulator’ object has no attribute ‘plan_SimAFRN’
Any suggestions??