LIF neuron has a resting potential 0. I was wondering how to make it -70mV without changing the code
Heading on over to the Nengo documentation for nengo.LIF, we see:
class nengo.LIF(tau_rc=0.02, tau_ref=0.002, min_voltage=0, amplitude=1):
...
min_voltage : float
Minimum value for the membrane voltage. If -np.inf, the voltage is never clipped.
In your model, the usage looks like so:
x = nengo.Ensemble(..., neuron_type=nengo.LIF(min_voltage=...)))
However, it is important to note that voltages in this model are unitless. That is, voltages $v(t)$ are normalized to the range $[0, 1]$ for convenience. To determine the resting potential that corresponds to -70mV you will need to do some math to determine its corresponding unitless value. Also note there is some interplay with the gains and biases of the neurons, and so care should be taken in interpreting these quantities in a biophysical context.
Hi,arvoelke
How is the voltage normalized?When V(t)=1, What is the corresponding voltage(mV)?
Thanks
There isn’t a well-defined mapping between the LIF’s unitless voltage and real biological voltage. This is so that we can avoid making assumptions about the types and magnitudes of inputs being provided to the neural network. Biological neurons have a pretty impressive dynamical range. If you wanted to define such a mapping, it is possible, but it would be specific to the model that you’re building. You would first have to determine the magnitude of the input currents driving the cells, and then you could use that value to figure out the appropriate scaling for the gain (which is, again, unitless for us) such that it introduces the expected change in voltage that results in a spike at V(t) = 1 and resets to a known reset value.
We have not had any situations (that I’m aware of) that have warranted that level of detail, so we don’t currently have any examples to show how you could do that mapping. But it is possible, and if someone was to do it, we could probably generalize that approach to make it more easily accessible.
Thank you very much for answering my question! By the way, I want to add a custom neuron model. Is there such an interface in nengo?
Yes, it is possible to add new neuron models to Nengo. We actually recently made the process of adding new neuron models to Nengo easier, but in order to use those changes you’ll have to use the development version of Nengo (see install instructions).
If you’re using the dev version, then follow this example. If you’re using Nengo 3.0.0, follow this example.
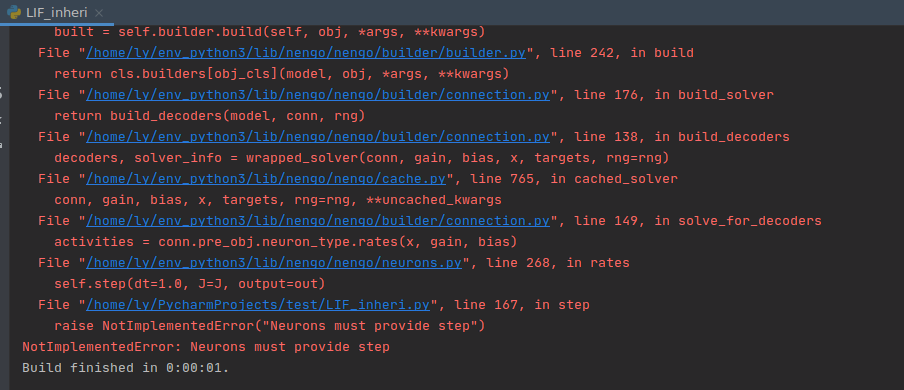
I tried to run the sample program of adding an object to nengo, but encountered the following problems:
Have you encountered this kind of problem? How to solve it?
Hm, it looks to me like you are using the developer installation of Nengo, but the Nengo 3.0 version of the example. In the developer installation, we renamed the step_method to step, which is why Nengo is looking for that function. If you have made some modifications to the example, you can try renaming your step_math function to step and seeing if that works (but there may also be some other modifications needed if you’re basing something off of the Nengo 3.0 version of the example). If you post the script that you’re working on that is raising the error I can provide some more specific advice.
Thank you very much for your answer!
Code show as below:
neuron_model.py (2.8 KB)
It looks like you were indeed using the Nengo 3.0 version of the example rather than the updated version. Try running this version, it should work – and if not, let me know what error you get.
neuron_model.py (1.2 KB)
Thank you for your detailed answer! By the way, I would like to ask, how to read the signal from the file as input?
To provide the contents of a file as input to the network, you would use whatever normal Python code you would like to read the file, then a nengo.Node to connect that to the rest of the network. Here’s a short example that reads a CSV file with two columns and provides that to the network.
The code achieves what I want. Thank you for your detailed and patient answers!