Hi,
I’m learning Nengo-DL and am stuck at a very basic thing. I want to create a network that predicts a number of signals.
I’m following the tutorials https://www.nengo.ai/nengo-dl/examples/tensorflow-models.html.
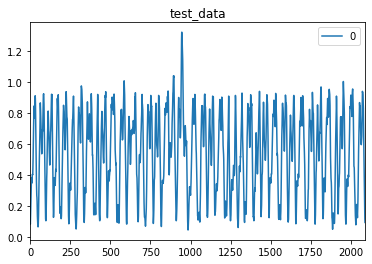
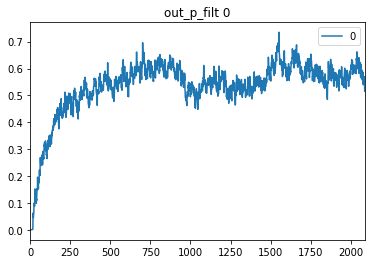
So I built a basic autoencoder model in Keras, which works. Then I changed it into Nengo Ensembles with RectifiedLinear neurons and it still works. But when I try to use spiking neurons the training looks successful and prints decreasing loss values, but I can’t find a way to get a prediction on test data. All I get is a constant value for all input dimensions.
My code looks like:
# train_data, validation_data, test_data are arrays [timesteps, features]
train_data = train_data[:, None, :]
validation_data=validation_data[:, None, :]
test_data = test_data[:, None, :]
earlyStopping= EarlyStopping(monitor='val_loss', patience=3, verbose=2, min_delta=1e-4, mode='auto')
lr_reduced = ReduceLROnPlateau(monitor='val_loss', factor=0.5, patience=1, verbose=2, min_delta=1e-4, mode='min')
with nengo.Network(seed=seed) as net:
# set up some default parameters to match the Keras defaults
net.config[nengo.Ensemble].gain = nengo.dists.Choice([1])
net.config[nengo.Ensemble].bias = nengo.dists.Choice([0])
net.config[nengo.Connection].synapse = None
net.config[nengo.Connection].transform = nengo_dl.dists.Glorot()
# # parameters for spiking neurons
net.config[nengo.Ensemble].max_rates = nengo.dists.Choice([100])
net.config[nengo.Ensemble].intercepts = nengo.dists.Choice([0])
net.config[nengo.Connection].synapse = None
inp = nengo.Node(output=np.ones(X1.values.shape[-1]))
#layers_sizes is a list of the layers' last dimension sizes
l = inp
for ls in layers_sizes:
hidden = nengo.Ensemble(100, ls
# ,neuron_type=nengo.RectifiedLinear()
,neuron_type=nengo.LIF(amplitude=0.01)
).neurons
nengo.Connection(l, hidden)
l = hidden
#the last layer without any activation
#I could not find a way to do it within nengo, so just adding a TF layer :-)
out = nengo_dl.Layer(
tf.keras.layers.Dense(ls))(l)
out_p = nengo.Probe(out, label="out_p")
with net:
nengo_dl.configure_settings(stateful=False, use_loop=False)
# when testing our network with spiking neurons we will need to run it
# over time, so we repeat the input/target data for a number of
# timesteps.
n_steps = 30
test_data = np.tile(test_data, (1, n_steps, 1))
sim = nengo_dl.Simulator(net, minibatch_size=32)
sim.compile(optimizer=tf.optimizers.Adam(lr=0.001),
loss='mean_squared_error')
sim.fit(train_data, train_data,
epochs=500,
batch_size=32,
shuffle=False,
callbacks = [earlyStopping, lr_reduced],
verbose = 2,
validation_data=(validation_data, validation_data))
preds = pd.DataFrame(data=sim.predict(test_data)[out_p][:,0,:])
preds.plot()
I’m struggling with this for a few days now, please help!