Hi,
I have a fairly basic question… I have the following model:
model = nengo.Network()
with model:
stim_teta1 = nengo.Node(Piecewise({0: 0, 1: 1.57}))
stim_teta2 = nengo.Node(Piecewise({0: 0, 2: 1.57}))
teta = nengo.Ensemble(n_neurons=600,
dimensions=2,
radius=np.sqrt(2 * 6.28**2),
seed = 0)
nengo.Connection(stim_teta1, teta[0])
nengo.Connection(stim_teta2, teta[1])
res_x = nengo.Ensemble(n_neurons=600,
dimensions=1,
radius=2,
seed = 0)
def calculate_x(teta):
**return np.cos(teta[0]) + np.cos(teta[1])**
nengo.Connection(teta, res_x, function=calculate_x)
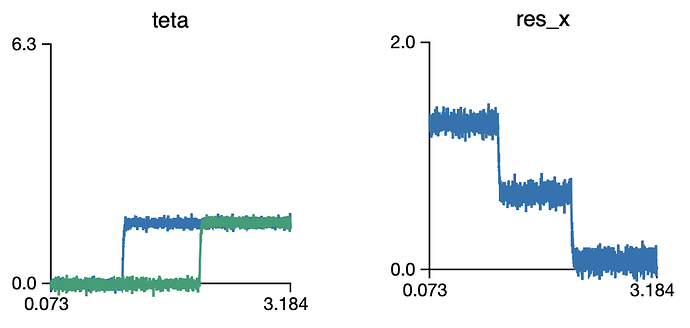
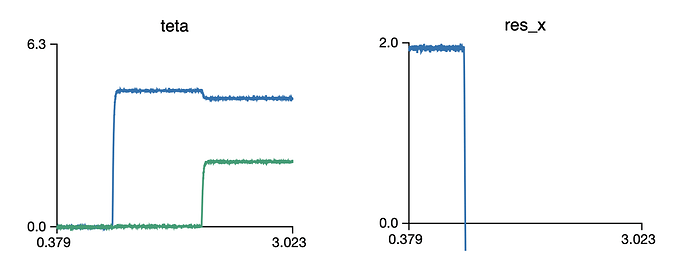
You would expect values of 2, 1, 0 in the res_x ensemble. However, I get the following:
I’m sure that there is an easy fix for it… and would appreciate your help…