Hi all,
I would like to implement differential Hebbian learning rules, described e.g. here: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6130884/. The general idea is to use the derivatives of pre- and post-synaptic neural activity in order to describe synaptic plasticity rules that capture higher-order dynamics. I have implemented multi-factor Hebbian plasticity rules in Nengo before, so I think I have a reasonable understanding of the relevant Nengo APIs. However, I wanted to ask the experts how they would go about differential plasticity rules. Would it be sufficient to keep copies of the pre- and post-synaptic activity vectors from the previous timestep, and use them with the current activity vectors to do simple forward integration? Or would it be better to keep a longer time history and use integration schemes that are more numerically stable? Any opinions are much appreciated!
Hi @iraikov and welcome back to the Nengo forums. 
I took a quick look the paper and the learning rule seems to use the instantaneous change in the pre and post synaptic activity, so it would probably work to just calculate this difference in the Nengo implementation. However, I can’t 100% say that this won’t cause instability issues… Unfortunately, customizing learning rules is a bit of black magic, and you really won’t know the full effects until you try it. So, I would recommend starting simple and just doing the difference calculation, and if that proves unstable, to slowly add complexity until it works the way you want it to.
Another thing you can do is filter your activities with a derivative filter, similar to how we filter the activities in the PES learning rule with a filter given by pes.pre_synapse.
Rather than using a pure derivative for your filter, though, I’d use a derivative combined with a lowpass filter. This acts the same as applying the lowpass filter to your signal, then taking the derivative, and will help stability.
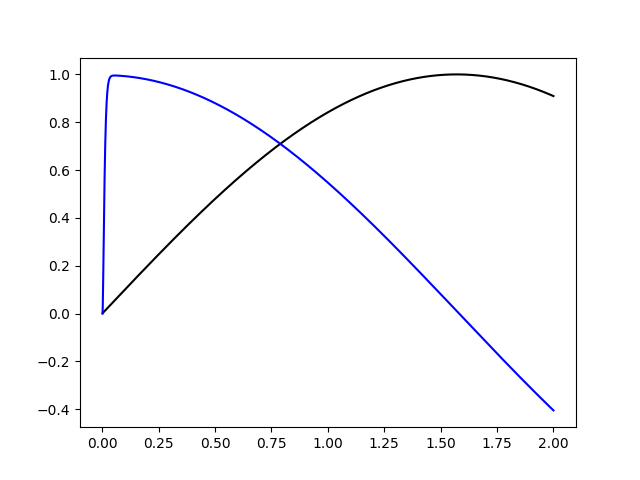
Here’s an example of such a filter applied to a sine wave:
import matplotlib.pyplot as plt
import nengo
import numpy as np
alpha_derivative = nengo.LinearFilter([1, 0], [1]).combine(nengo.Alpha(0.005))
with nengo.Network() as net:
u = nengo.Node(lambda t: np.sin(t))
up = nengo.Probe(u)
udp = nengo.Probe(u, synapse=alpha_derivative)
with nengo.Simulator(net) as sim:
sim.run(2.0)
plt.plot(sim.trange(), sim.data[up], "k")
plt.plot(sim.trange(), sim.data[udp], "b")
plt.show()
Thank you, this is very helpful!
If I try to create two LinearFilters (one for the pre-synaptic and one for the post-synaptic activity), I get the following error:
nengo.exceptions.ValidationError: obj: Can only combine with other LinearFilters
What am I doing wrong?
What are you trying to combine with? It sounds like you’re trying to combine with something that is not a LinearFilter. Check the type of the argument to combine.
(You can check if an object obj is a LinearFilter with isinstance(obj, nengo.LinearFilter).)
Sorry I was not clear. I just tried to combine with Lowpass.
Can you post an example, along with the Nengo version you’re using? You should be able to combine with a Lowpass instance (since Lowpass is a subclass of LinearFilter).
My mistake was trying to create the LinearFilter instance in the constructor of a subclass of LearningRuleType. The LinearFilter.combine method was actually getting a SynapseParam. Sorry about the noise and thanks a lot for your help!