Hey all,
I am running in some problems with a nengo implementation of a model executing a delayed-response task. I have two ensembles (lets call them A and B) that both represent an orientation. Ensemble A represents the orientation by means of sine and cosine of theta. Ensemble B represents the orientation by means of sine and cosine of theta multiplied by some unkown constant a that varies with time.
So we have the following:
ensemble A represents: sine(theta), cosine(theta)
ensemble B represents: a sine (theta), a cosine(theta)
I need to decode the difference between theta represented in ensemble A and B, For this I could simply use the arctan2 function:
diff_thetaAB= arctan2(sinA, cosA)-arctan2(sinB, cosB)
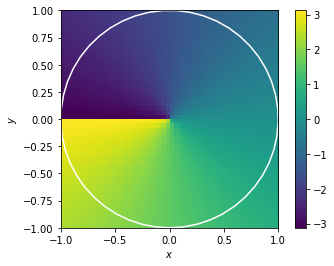
which should also gets rid of the unkown mupltiplier a as arctan2(sin, cos) equals arctan2(a sin, a cos)
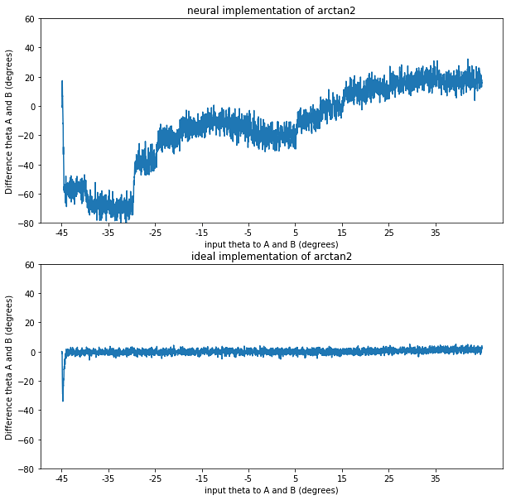
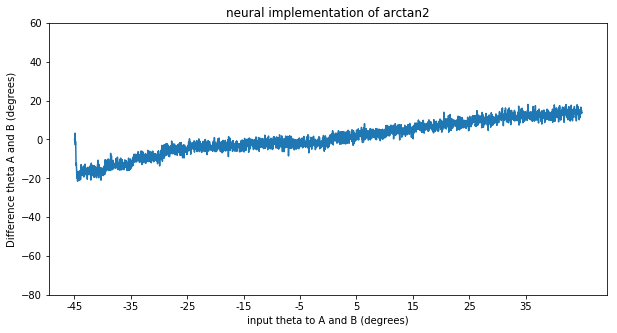
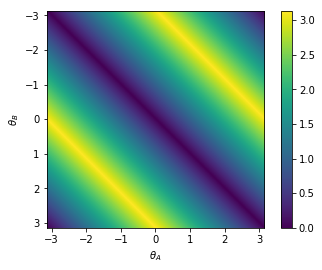
This all works fine if I use a non-neural implementation of arctan2 (by setting the neuron_type to nengo.Direct()). However as soon as I calculate the arctan2 function with spiking neurons I start to run into problems. I’ve added a simple model that just highlights the problem below. The neurons do quite a bad job of approximating the arctan2 function (which might be expected as its shape is not continuous). However they don’t just compute a noisy version of arctan2, they are consistently worse for some theta as shown in the second graph below.
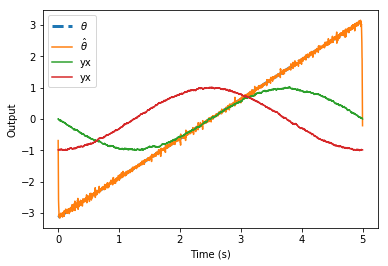
The model I used to create the graph has two ensembles A and B that are both fed the same orientation, with a set to 0.1. The input goes from theta -45 to theta is 45 degrees in steps of 5 degrees every 0.25 seconds as noted on the x axis. The y-axis represents the difference in theta decoded from the sine and cosine of ensemble A and B. First with a neural implementation and secondly with a direct implementation.
As seen in the direct implementation we would expect a difference of 0, as both ensemble A and B are fed the same orientation. However the neural implementation consistently gives a “negative” difference for a negative orientation and a “positive” difference for a positive input orientation as can be seen in the graph. (It wouldn’t be problematic for my model if the neurons just did a noisy implementation, but that is clearly not the case)
So my question is, why is there this consistent error in the neural implementation of the arctan2 function? Does this have something to do with the shape of the function? If so, is it possible to implement it in such a way that gets rid of this problem? (by optimizing it just over the range for which it is continuous maybe?) Or is there another way I could decode the orientation difference theta represented in two ensembles from their sine and cosine of theta?
Please let me know if you have any questions. I would be very grateful if anybody can help me shed some light on this problem!