Greetings,
I am using SPA to build a biologically plausible knowledge representation network but I have some questions.
1st Question
First of all I am using an autoassociative thresholding memory to store pointers of the form
A = C1 * K1 + C2 * N1;
B = C1 * (K2 + N2) + C2 * K3;
which I query as following:
vocab['A'] * ~ vocab['C1'] >> model.memory
Every time I rerun my query though I get a different result. Usually it is the correct one but once in a while it can be wrong. In the case of using the second pointer B, which must return the sum (K2 + N2), the errors are more frequent.
If I can hazard a guess I would say that this is due to the fact that semantic pointers get instantiated randomly so that is why every time I get different results, but why do I get errors too?
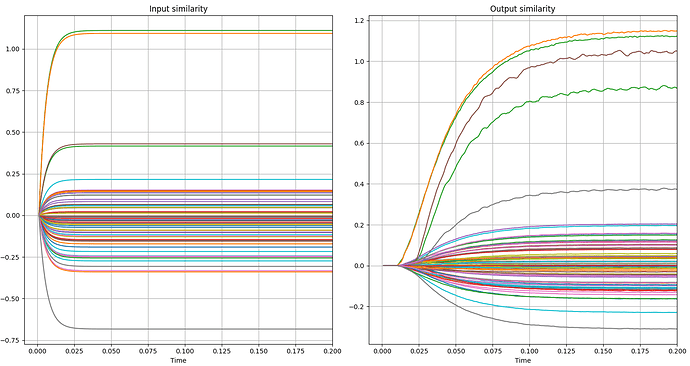
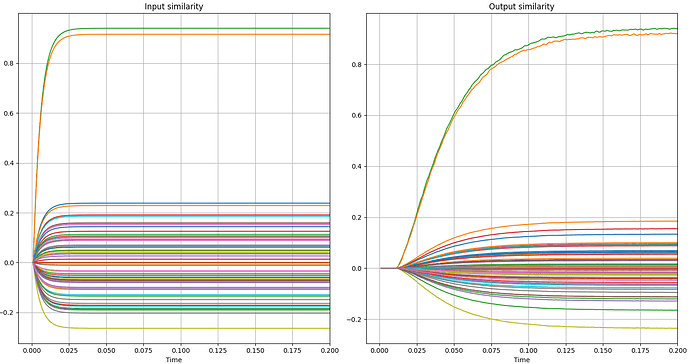
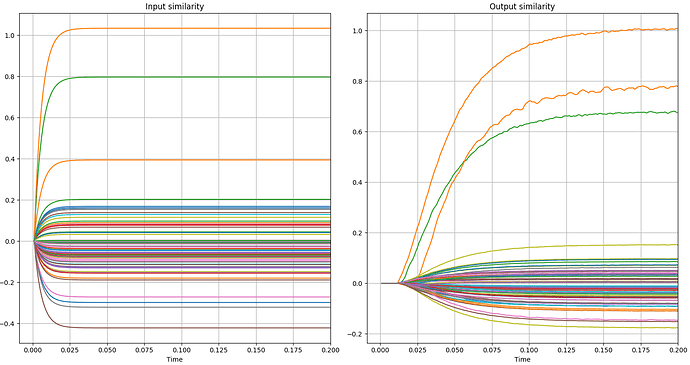
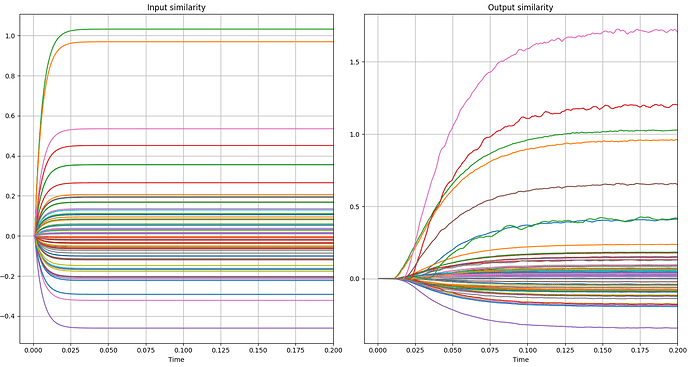
An example can be seen in the following graphs. In the first two you can see the variation I am talking about and in the next two you can see errors, as the correct green and orange lines are mistakenly lower in the similarity measurements.
2nd Question
When I query the memory like this
vocab['A'] * ~ vocab['C1'] >> model.memory
the model runs very fast, but when I tried the query like this
first = spa.Transcode('A', output_vocab=terms)
second = spa.Transcode('C1', output_vocab=terms)
term = spa.State(vocab, neurons_per_dimension = 5)
query = spa.State(vocab, neurons_per_dimension = 5)
nengo.Connection(first.output, term.input)
nengo.Connection(second.output, query.input)
term * ~query >> model.memory
the model runs incredibly slow. Why? What is the difference?
3rd Question
When using standard Nengo you can choose the type of neurons and synapses you use but I can’t find any option on how to see and also change what kind of neurons and synapses I use when I model with the SPA. What is the default neuron types used in SPA?