I am utilizing a Nengo network to generate an updated activation array to control a robotic arm.
Given the current robot arm position as a (n,d) array, I want to learn the (10, ) activation array to reach some certain target position.
This is a general idea of the connections I have (without the PES learning):
input_node -> input_layer-> output_layer -> output_node
input_node: Represents the current arm position. The shape = (n, d) numpy array representing the current arm position. d = dimension (x, y, z) and n = positions along a dimension
input_layer: The spiking neurons to represent the input
output_layer: The spiking neurons to represent the output. This is of shape (10, ), representing the activation required. Each element has to be between [-1.0, and 1.0].
output_node: The decoded output from the spiking output_layer
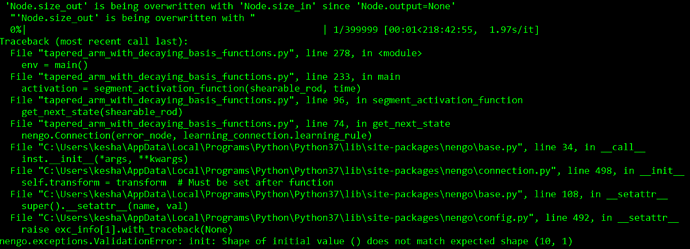
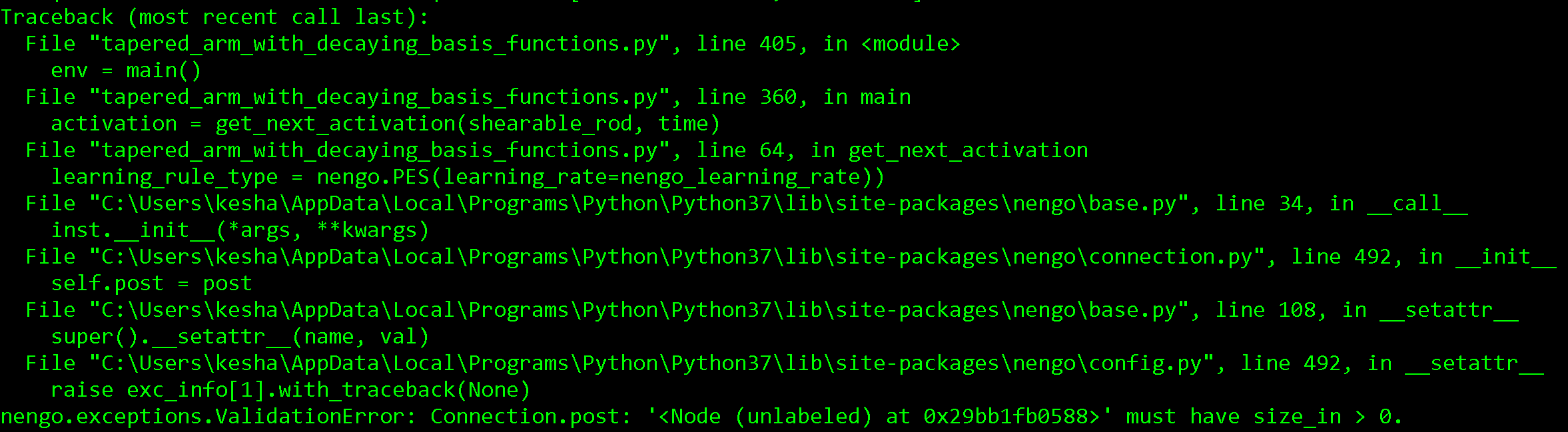
The trouble comes when I connect the input_layer to the output_layer with the PES learning rule. My error signal is simply the distance of the end of the robot arm to the target position, but this does not match the dimensionality of the ensembles in the connection.
Below is the code:
global most_recent_activation
global learned_weights
most_recent_position = np.array(shearable_rod.position_collection)
n = most_recent_position.shape[0]
d = most_recent_position.shape[1]
# Define the nengo network
with nengo.Network() as net:
# Returns the current arm position
def get_rod_position(t):
current_position = np.array(shearable_rod.position_collection)
return current_position.ravel()
# Calculates the distance between the end of the arm and the target
def reward(t):
end_position = most_recent_position[:,-1]
distance = np.linalg.norm(end_position - target_position)
#print("distance: ", distance)
return -distance
# Spiking neurons representing input rod position: get_rod_position() -> rod_posistion_node -> rod_position_ensemble
input_node = nengo.Node(get_rod_position)
input_layer = nengo.Ensemble(n*d, dimensions = n*d)
# Spiking Neurons representing hidden layer
#hidden_layer = nengo.Ensemble(n_neurons = int((2/3)*(n*d) + 13*2), dimensions = n*d)
# Spiking neurons representing output layer
output_layer = nengo.Ensemble(n_neurons = 128, dimensions = 10,
intercepts = Uniform(low=-1.0, high=1.0))
output_node = nengo.Node(size_in = n*d, size_out = 10) # decodes the output of the spiking neurons
# Weights to transform from input to output_layer
weights = np.random.normal(size=(10, n*d))
if (learned_weights is not None):
weights = learned_weights
# Conncetions: input_node -> input_layer -> hidden_layer -> output_layer -> output_node
nengo.Connection(input_node, input_layer)
learning_connection = nengo.Connection(input_layer, output_layer,
transform = weights,
learning_rule_type = nengo.PES(learning_rate=3e-4))
#WeightSaver(learning_connection, "learning_connection_weights")
# W2 = np.random.normal(size=(13*2, n*d))
error_node = nengo.Node(output = reward, size_out = 1)
nengo.Connection(error_node, learning_connection.learning_rule)
nengo.Connection(output_layer, output_node)
net.probe_output = nengo.Probe(output_node)
The error I get is:
My questions are as follows:
-
How can I connect the input_layer to the output_layer and learn with the scalar error signal? This is the only notion of error that I have, I can not use a pre - post error signal since I don’t know the true (10, ) activation array.
-
Is input_node -> input_layer-> output_layer -> output_node connection approach correct? The only reason I convert input_node to input_layer is because I am unable to connect the node to the ensemble with the PES learning rule. It seems to require two ensembles.
-
Do I need to have output_layer -> output_node to decode the output of the spiking neural network?
-
Later I will need to save the weights learned as I will be running this network in an online fashion. How can I do this? I could not find any good example or tutorials online.
Thank you!