This is correct.
You are correct in the observation that currently, all of the transformations (convolution, average pooling, etc.) supported by the NengoDL converter are linear transformations. The reason for this is somewhat nuanced, however.
In my previous post, I mentioned that Nengo and NengoLoihi networks can “compute” (approximate) any function, even non-linear functions. The obvious question here is, of course, why only linear transformations are included in the NengoDL converter by default.
Let us examine linear transformations first. Since the transformations are linear, they can be incorporated into the connection weights in a straightforward manner using the appropriate matrix operations. These matrix operations are defined by the linear transformations themselves and have no addition parameters to tweak (since it’s just a matrix multiply), so we know the exact “solution” as to how to integrate such things into the Nengo network. Since we know the exact “solution”, linear transformations can be implemented in the NengoDL converter with too much difficulty, and are included by default.
What about non-linear functions? In Nengo, non-linear functions are “computed” by solving for the appropriate connection weights such that for a given input to a neural ensemble, the weighted activation functions approximate the desired function. Note, however, that in all of our Nengo examples that compute non-linear functions (e.g., product, square, etc.), an ensemble of neurons is required to compute the function. This is where the difficulty lies in incorporating such functionality into an automatic converter. In TF for example, you could apply a max pooling function on the output of a layer, like so:
input --> neurons --> max pooling --> output
But, to do the same thing in Nengo, since the max pooling function is non-linear, you’ll need an additional ensemble of neurons to perform this computation:
input --> neurons --> ensemble --> (weights computing max pooling) --> output
The specific details of this additional ensemble are typically application / user dependent (e.g. what range of values the input has, to what accuracy does the user want the non-linear function approximated, number of neurons to use in this additional ensemble, neuron parameters for this ensemble, etc.), so it becomes impossible to build a “default” converter for these non-linear functions. Instead, we leave it up to the user to extend the NengoDL converter to implement the desired non-linear functions to their own specifications.
Just to summarize, this statement is partially correct, and partially incorrect. As I described above, something like max pooling isn’t natively supported by the NengoDL converter, so in that sense it “cannot be converted to a Nengo object”. However, the function itself can be approximated in a Nengo network, so if the user takes the time to implement their own converter function to do so, something like max pooling can be converted to use only Nengo objects.
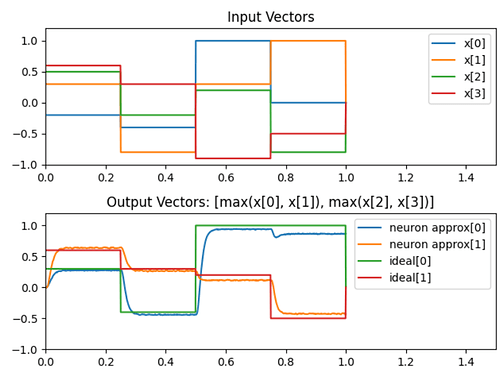
Here’s some code demonstrating the max pooling function being computed using only Nengo objects: test_max_pool.py (1.3 KB)
The max pooling operation is being performed on a 4D vector, where the function computes [max(x0, x1), max(x2, x3)]. And this is what the output graph looks like:
Note: I made the ensemble with 1000 neurons because I didn’t want to fuss around with optimizing it. I just knew that 1000 neurons would have been more than plenty to approximate this function. Even 200-ish will work, but you can test that on you own.
Note 2: There are also other tricks you can use to optimize the neural implementation of the max pooling operation. As an example, instead of having 1 ensemble to the full 4D input, I could have used an EnsembleArray (source docs here) and split the input into 2 sub-ensembles before doing the max function. This approach scales up better to larger inputs. E.g., if you had a 16x16 matrix and you wanted to do a 2x2 max pooling, instead of a 256D ensemble with something like 25600 neurons (which would take a long time to solve the decoders for), you’ll instead use an EnsembleArray with 128 sub-ensembles, with each sub-ensembles being 4D and maybe 400 neurons (which doesn’t take a long time to solve the decoders). i.e., instead of this:
nengo.Ensemble(25600, 256)
we do this:
nengo.networks.EnsembleArray(400, n_ensembles=128, ens_dimensions=4)