Hello @xchoo, thanks for giving it second thoughts. Below is a custom script I wrote for the calculation of ISI (haven’t implemented the exact function yet, but it should be straightforward, given that I am able to access the spikes (with amplitude 1/dt) in my _get_isi()). My goal is to calculate ISI for more than 1 input scalar (i.e. more than one inp_node1).
SEED = 89
x1, x2 = 0.9, 0.1
def _get_isi(t, x):
print(x)
#return x[0], x[1]
with nengo.Network(seed=SEED) as net:
# Create Input Nodes.
inp_node1 = nengo.Node(x1)
inp_node2 = nengo.Node(x2)
# Create 2 Ensembles.
ens1 = nengo.Ensemble(n_neurons=1, dimensions=1, seed=SEED, radius=1)
ens2 = nengo.Ensemble(n_neurons=1, dimensions=1, seed=SEED, radius=1)
# Create the ISI Node.
isi_node = nengo.Node(output=_get_isi, size_in=1)
# Connect the Input nodes to the Input ensembles.
nengo.Connection(inp_node1, ens1, synapse=None) # Default Synapse is Lowpass(0.005)
nengo.Connection(inp_node2, ens2, synapse=None) # Default Synapse is Lowpass(0.005)
# Connect the Input ensembles to the ISI node.
nengo.Connection(ens1.neurons, isi_node, synapse=None)
nengo.Connection(ens2.neurons, isi_node, synapse=None)
# Check the representation of inputs.
probe1 = nengo.Probe(ens1, synapse=nengo.Lowpass(0.005))
probe2 = nengo.Probe(ens2, synapse=nengo.Lowpass(0.005))
# Check the spiking pattern.
spikes1 = nengo.Probe(ens1.neurons) # Default synpase is None
spikes2 = nengo.Probe(ens2.neurons) # Default synpase is None
with nengo.Simulator(net) as sim:
sim.run(1)
vctr_points_1, activities1 = tuning_curves(ens1, sim)
vctr_points_2, activities2 = tuning_curves(ens2, sim)
It runs error free when size_in=1 i.e. when I access spikes for only one input (although there are two connections made to the isi_node:
# Connect the Input ensembles to the ISI node.
nengo.Connection(ens1.neurons, isi_node, synapse=None)
nengo.Connection(ens2.neurons, isi_node, synapse=None)
which I thought… it should throw an error due to size_in=1, but no… no error is thrown. However, when I set size_in=2 to access the spikes for both inputs (i.e. inp_node1 and inp_node2) in function _get_isi(), it throws this error: ValidationError: Connection.transform: Transform output size (1) not equal to connection output size (2). I looked through few scant examples of creating and using Node with size_in=2 (or more), and found that one needs to mention a transform=np.eye(2) in the nengo.Connection() while creating a connection between the ensemble and the isi_node, but that gave me an expression of creating a multi-dimensional ensemble - which I guess is not useful for me, as I want to record ISI from each individual neuron (as done in Nengo-DL, e.g. the vector input is presented to the first Conv/Dense layer for n_steps and each neuron in the Ensemble represents one scalar element of the vector input). Can you please help me with recording the ISI from individual neurons, each representing a scalar?
I have few more questions though, which are sort of related to my question here. In the following tuning curve plots:
plt.plot(vctr_points_1, activities1)

plt.plot(vctr_points_2, activities2)

you can see that both the neurons have same curve, and that’s because of the same seed value. I see that a scalar of value 0.7 or more will lead both the neurons to spike, and that indeed is the case as can be seen below (filtered output for scalar 0.9):
plt.plot(sim.data[probe1])
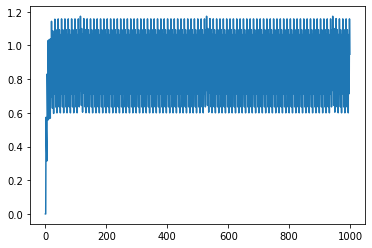
However, for scalar input of 0.1, the neuron is not supposed to spike as per the tuning curve plot, and that indeed is reflected below in the filtered output plot.
plt.plot(sim.data[probe2])
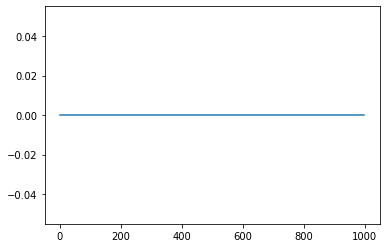
Now, since the radius is set to one, I am supposed to input values in range [-1, 1] only, but as can be seen above, it’s not guaranteed that a single neuron will spike for that input (a group of neurons will do). So how is it done in Nengo-DL where one neuron represents each scalar input (either direct input or calculated ones in the network e.g. Convolved ones). Please let me know this too.