Hey,
I am trying to work on a converter example and have came across the issue
nengo.exceptions.SimulationError: Number of saved parameters in lidar_params (25) != number of variables in the model (24)
but this only happens when I try to add synapses when they are none it works fine
modified from the dewolf blog – Keras to SNN example
Just to add, found that the flatten layer seem to get split into 2 in example where synapse = none and with synapses it condenses into one layer in the weights of the tensor graph
inp = Input(shape=(7999, 1), batch_size=minibatch_size)
conv1 = Conv1D(64, 3, activation='relu', padding='same', kernel_initializer='he_normal')(inp)
conv1 = Conv1D(64, 3, activation='relu', padding='same', kernel_initializer='he_normal')(conv1)
pool1 = MaxPooling1D(pool_size=(2))(conv1)
conv2 = Conv1D(128, 3, activation='relu', padding='same', kernel_initializer='he_normal')(pool1)
conv2 = Conv1D(128, 3, activation='relu', padding='same', kernel_initializer='he_normal')(conv2)
pool2 = MaxPooling1D(pool_size=(2))(conv2)
conv3 = Conv1D(256, 3, activation='relu', padding='same', kernel_initializer='he_normal')(pool2)
conv3 = Conv1D(256, 3, activation='relu', padding='same', kernel_initializer='he_normal')(conv3)
pool3 = MaxPooling1D(pool_size=(2))(conv3)
conv4 = Conv1D(512, 3, activation='relu', padding='same', kernel_initializer='he_normal')(pool3)
conv4 = Conv1D(512, 3, activation='relu', padding='same', kernel_initializer='he_normal')(conv4)
# drop4 = Dropout(0.5)(conv4)
pool4 = MaxPooling1D(pool_size=(2))(conv4)
conv5 = Conv1D(1024, 3, activation='relu', padding='same', kernel_initializer='he_normal')(pool4)
conv5 = Conv1D(1024, 3, activation='relu', padding='same', kernel_initializer='he_normal')(conv5)
# drop5 = Dropout(0.5)(conv5)
flat = Flatten()(conv5)
dense = Dense(units=128)(flat)
dense = Dense(units=1024)(dense)
out = Dense(units=4096)(dense)
model = tf.keras.Model(inputs=inp, outputs=out)
converter = nengo_dl.Converter(
model,
swap_activations={tf.nn.relu: nengo.RectifiedLinear()},
max_to_avg_pool=True)
net = converter.net
nengo_input = converter.inputs[inp]
nengo_output = converter.outputs[out]
nengo_conv1 = converter.layers[conv1]
with converter.net as net:
probe_conv5 = nengo.Probe(nengo_conv1, label='probe_conv5')
# run training
adam = tf.keras.optimizers.Adam(learning_rate=0.0001, beta_1=0.9, beta_2=0.999, amsgrad=False)
with nengo_dl.Simulator(net, minibatch_size=minibatch_size, seed=0) as sim:
sim.compile(
optimizer=adam,
loss={nengo_output: "mse"},
)
sim.fit(train_histograms, {nengo_output: train_images}, epochs=30)
# save the parameters to file
sim.save_params("lidar_params")
# test gain scaling with non-spiking neurons
def run_network(activation, params_file, gain_scale=1, synapse=None):
converter = nengo_dl.Converter(
model,
swap_activations={tf.nn.relu: activation},
scale_firing_rates=gain_scale, max_to_avg_pool=True,
)
net = converter.net
nengo_input = converter.inputs[inp]
nengo_output = converter.outputs[out]
nengo_conv5 = converter.layers[conv5]
with net:
probe_conv5 = nengo.Probe(nengo_conv5)
if synapse is not None:
for conn in net.all_connections:
conn.synapse = synapse
n_test = 6
with nengo_dl.Simulator(net, minibatch_size=1, seed=0) as sim:
sim.load_params(params_file)
data = sim.predict({nengo_input: test_histograms[:n_test]})
# plot network predictions
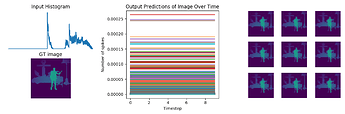
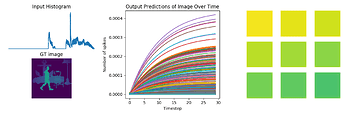
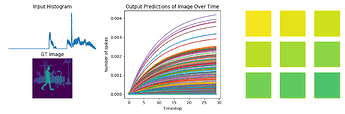
fig = plt.figure(figsize=(12, 10))
fig.suptitle(number)
for test in range(n_test):
plt.subplot(3, 2, test+1)
plt.imshow(data[nengo_output][test, 0].reshape((64, 64)), cmap='viridis_r', vmin=0, vmax=1) # cmap=plt.cm.binary)
plt.axis("off")
# plt.figure()
# plt.plot(test_labels[:n_test].squeeze(), 'rx', mew=15)
# plt.imshow(data[nengo_output][].reshape(64, 64), cmap='viridis_r', vmin=0.02, vmax=1) # plt.ylabel('Digit')
# plt.xlabel('Batch')
# plt.ylim([-.5, 9.5])
# plt.xticks(range(5), range(5))
# plt.yticks(range(10), range(10))
plt.title('Network predictions for {}'.format(number)) ### change to showing image
# plot neural activity
plt.figure()
n_neurons = 5000
activity = data[probe_conv5]
if activation == nengo.SpikingRectifiedLinear():
activity = activity * 0.001 * gain_scale
print('Max value: ', np.max(activity.flatten()))
plt.plot(activity[:, :, :n_neurons].reshape(-1, n_neurons))
plt.title('Neuron activity for {}'.format(number))
# plot raw network output (the first 5 dimensions)
plt.figure()
for ii in range(5):
plt.subplot(5, 1, ii + 1)
plt.plot(data[nengo_output][0].reshape(-1, 10)[:, ii])
plt.ylabel('%i' % ii)
plt.tight_layout()
plt.title('Network output for {}'.format(number))
number = 1
# run the network in rate mode
run_network(
activation=nengo.RectifiedLinear(),
params_file='lidar_params')
number = 2
# run the network with spiking neurons
run_network(
activation=nengo.SpikingRectifiedLinear(),
params_file='lidar_params',
)
number = 3
# run the network with spiking neurons showing each input for 100ms
train_images, train_histograms, test_images, test_histograms = load_lidar(plot=False, n_steps=100)
run_network(
activation=nengo.SpikingRectifiedLinear(),
params_file='lidar_params',
)
number = 4
# run the network with spiking neurons showing each input for 100ms and using synapses
for synapse in [0.001, 0.005, 0.01]:
run_network(
activation=nengo.SpikingRectifiedLinear(),
params_file='lidar_params',
synapse=synapse,
)
number = 5
# test gain scaling with spiking neurons showing each input for 30ms
train_images, train_histograms, test_images, test_histograms = load_lidar(plot=False, n_steps=30)
for gain_scale in [1, 10, 20]:
run_network(
activation=nengo.SpikingRectifiedLinear(),
params_file='lidar_params',
gain_scale=gain_scale
)