Hi! I have a hardware device that creates Poisson Spikes but its tuning curve is a Rectified Exponential as shown in Fig 1. I want to use Nengo to simulate many of my devices as a neuron population. I have tried to create my own rectified exponential neuron in Nengo by modifying the RectifiedLinear neuron class as shown in the code snippet “Modified RectifiedLinear Neuron type class” and it performs with some limited success. The code in Nengo for a single device is also shown. This implementation doesn’t work well with Nengo’s random initialization of neurons, which is expected since Nengo is still initializing for a RectifiedLinear Neuron. I was wondering if anyone has some tips for implementing a custom neurons in Nengo. If any part of my post is not clear, please reach out to me. Thank you!
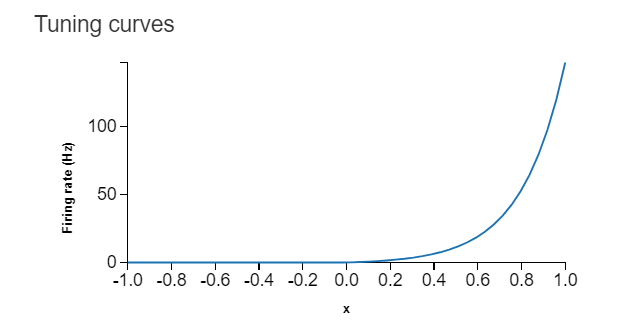
Fig 1
Nengo Code
import nengo
model = nengo.Network()
with model:
stim0 = nengo.Node([0])
max_rate_mat = [5,5]
encoder_mat = [[1],[-1]]
intercept_mat = [0.0,0.0]
a = nengo.Ensemble(n_neurons=2, dimensions=1, max_rates=max_rate_mat, encoders=encoder_mat, intercepts=intercept_mat,
neuron_type=nengo.PoissonSpiking(nengo.RectifiedLinear()))
nengo.Connection(stim0, a)
Modified RectifiedLinear Neuron type class
class RectifiedLinear(NeuronType):
"""A rectified linear neuron model.
Each neuron is modeled as a rectified line. That is, the neuron's activity
scales linearly with current, unless it passes below zero, at which point
the neural activity will stay at zero.
Parameters
----------
amplitude : float
Scaling factor on the neuron output. Corresponds to the relative
amplitude of the output of the neuron.
initial_state : {str: Distribution or array_like}
Mapping from state variables names to their desired initial value.
These values will override the defaults set in the class's state attribute.
"""
negative = False
amplitude = NumberParam("amplitude", low=0, low_open=True)
def __init__(self, amplitude=1, initial_state=None):
super().__init__(initial_state)
self.amplitude = amplitude
def gain_bias(self, max_rates, intercepts):
"""Determine gain and bias by shifting and scaling the lines."""
max_rates = np.array(max_rates, dtype=float, copy=False, ndmin=1)
intercepts = np.array(intercepts, dtype=float, copy=False, ndmin=1)
gain = max_rates / (1 - intercepts)
bias = -intercepts * gain
return gain, bias
def max_rates_intercepts(self, gain, bias):
"""Compute the inverse of gain_bias."""
intercepts = -bias / gain
max_rates = (1 - intercepts)
return max_rates, intercepts
def step(self, dt, J, output):
"""Implement the rectification nonlinearity."""
output[...] = self.amplitude * (np.exp((np.maximum(0.0, J)))-1)