Hi,
I’m looking at the tutorial of NEF algorithm (https://www.nengo.ai/nengo/examples/advanced/nef-algorithm.html) and also playing with the intercepts.
I can understand and also derive how the gain and bias are obtained in generate_gain_and_bias function:
def generate_gain_and_bias(count, intercept_low, intercept_high, rate_low, rate_high):
gain = []
bias = []
for i in range(count):
intercept = random.uniform(intercept_low, intercept_high)
z = 1.0 / (1 - math.exp((t_ref - (1.0 / rate)) / t_rc))
g = (1 - z) / (intercept - 1.0)
b = 1 - g * intercept
gain.append(g)
bias.append(b)
return gain, bias
I slightly modified the above function in order to get evenly distributed intercepts, so it becomes:
def generate_gain_and_bias(count, intercept_low, intercept_high, rate_low, rate_high):
gain = []
bias = []
##################### my modification #####################
intercepts = numpy.linspace(intercept_low, intercept_high, count)
for i in range(count):
##################### my modification #####################
intercept = intercepts[i]
z = 1.0 / (1 - math.exp((t_ref - (1.0 / rate)) / t_rc))
g = (1 - z) / (intercept - 1.0)
b = 1 - g * intercept
gain.append(g)
bias.append(b)
return gain, bias
However, the actual tuning is shown below. The intercepts are not evenly distributed. Though it looks more ‘even’ than the original version (the uniform distributed intercept).
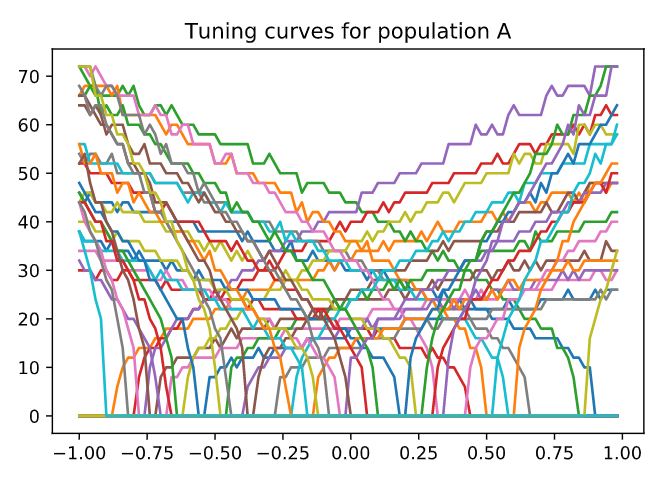
In addition, there may be a bug, just right below the step 2 section
v_A = [0.0] * N_A # voltage for population A
ref_A = [0.0] * N_A # refractory period for population A
input_A = [0.0] * N_A # input for population A
The refractory period of A and B are set to 0, but when calculate gain and bias, t_ref = 0.002 are used. First I suspect this may cause the mismatch between the desired and actual intercept. I change ```
ref_A and ref_B to 0.002, but it doesn’t fix the problem. desired and actual intercept still mismatch.
Then I checked how the intercept of LIF neuron is calculated in nengo source code. but the function is not implemented, so I guess nengo.neurons.NeuronType.gain_bias will be called to obtain the gain and bias. This function does not calculate gain and bias analytically, it looks like monte carlo method.
So my question is:
I believe the analytical solution (shown in first code block) to calculate gain and bias of LIF neuron is correct. But the actual result mismatches with my expectation, thet actual intercepts are not really evenly distributed. So I don’t know what’s going wrong.
My result should be quite easy to reproduce. The only relevant modification is shown in the second block. You can also set ref_A and ref_B to 0.002 instead of 0, but this doesn’t affect the tuning curve visibly.