Hello! I am working on the tutorial 20: spa_action and I was wondering if it is possible to delete the connection from the state’s input (input from vision) to Striatum D2 and STN in basal ganglia to show a dysfunction of indirect pathway?
Hi @cansukucuksozen, and welcome to the Nengo forums! ![]()
Yes, it is! But it requires a bit of understanding of the internals of how the basal ganglia network is constructed in Nengo. To modify a connection in a Nengo network, the basic premise is as follows:
- Iterate through all of the connections in the network.
- For each connection in the iteration, identify the connection you want to modify by checking the connection’s
pre_objandpost_objattributes. Thepre_objis the connection’s origin, andpost_objis the connection’s destination.
In Python, this would be:
for conn in model.all_connections:
if conn.pre_obj is <target_source> and conn.post_obj is <target_dest>:
<do stuff>
With this basic code, lets see how to apply it to the BG connections you want to modify.
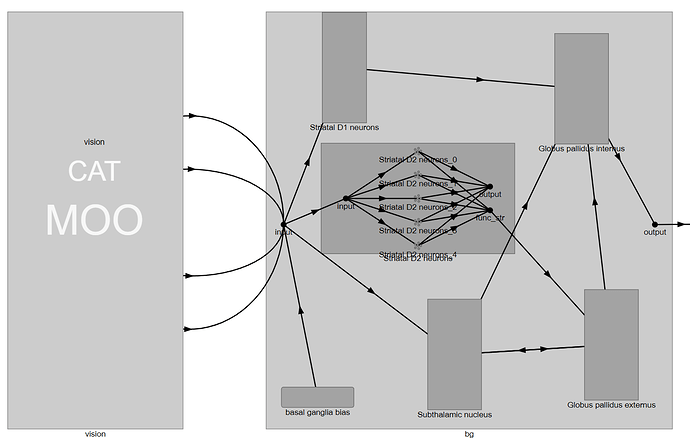
First, we need to identify the pre_obj and post_obj objects of the connections you want to modify. If you expand the BG network in NengoGUI (double click the network box), you’ll see something like this:
So, this tells us that the vision input gets routed to the BG input node, the from there, gets connected to the individual BG ensembles (STN, strD1, strD2, etc.). Inside each of these ensembles, it gets connected to another input node. From this, and from the BG network source code, we can determine that the connections we want to modify have a pre_obj of model.bg.input, and a post_obj of either model.bg.strD2.input or model.bg.stn.input.
Next we have to modify the connection attributes to lesion just the ones we want. This part is where it gets a little complicated. By default, the connections in the BG have a Dense transform type. We’ll need to make a new transform type with the values we want (with the lesioned bits).
We can get the default values for the connection transform with:
transform = numpy.eye(conn.transform.size_in) * conn.transform.sample()
Next, looking at the SPA actions, only the first 4 have connections to vision, so to lesion just those values, we’ll set the first four rows of the transform matrix to 0:
transform[0:4, :] = 0
Note: You can do a “partial” lesion by choosing a new value that is not fully 0. As an example, this will halve the transform’s initial value:
transform[0:4, :] *= 0.5
Finally, we’ll replace the connection’s transform with the new one:
conn.transform = nengo.transforms.Dense(conn.transform.shape, transform)
**Note: If you want to lesion the connections entirely, you can just do this, without the fancy indexing stuff:
conn.transform = nengo.transforms.Dense(conn.transform.shape, 0)
Putting all of this together, we have the code that will perform the lesioning for us:
for conn in model.all_connections:
if conn.pre_obj is model.bg.input and (
conn.post_obj is model.bg.stn.input or conn.post_obj is model.bg.strD2.input
):
transform = np.eye(conn.transform.size_in) * conn.transform.sample()
transform[0:4, :] = 0 # Perform lesion
conn.transform = nengo.transforms.Dense(conn.transform.shape, transform)
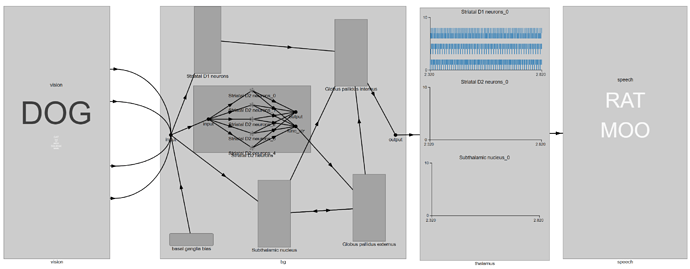
Now, let’s see if this had any effect. If we run the network with a “DOG” input, we see something like this:
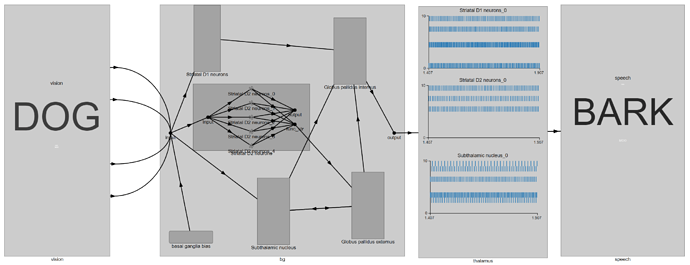
We observe that with a “DOG” input, there is no spiking activity in the StrD2 and STN populations. If we disable the lesioning code, this is what we see instead:
Note: If the images are a little hard to read, right click on the image and select “open in new tab”.