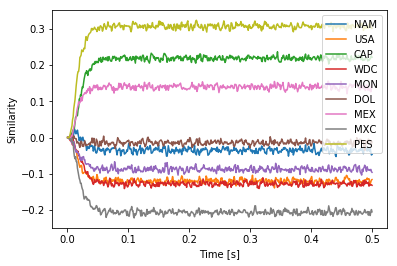
I believe the fundamental issue is the above vector algebra assumes that each vector is its own inverse (i.e., v == ~v, or v * v == 1, where 1 is the identity vector).
For example, when considering:
$$\begin{align}
F_{UM} &= \text{USTATES} \ast \text{MEXICO} \\
&= \left[ (\text{NAM} \ast \text{USA}) + (\text{CAP} \ast \text{WDC}) + (\text{MON} \ast \text{DOL}) \right] \ast \left[ (\text{NAM} \ast \text{MEX}) + (\text{CAP} \ast \text{MXC}) + (\text{MON} \ast \text{PES}) \right] \\
&\overset{?}{=} \left[ (\text{USA} \ast \text{MEX}) + (\text{WDC} \ast \text{MXC}) + (\text{DOL} \ast \text{PES}) + \text{noise} \right]
\end{align}$$
the question of equality depends on whether $(\text{NAM} \ast \text{USA}) \ast (\text{NAM} \ast \text{MEX}) \overset{?}{=} \text{USA} \ast \text{MEX}$, which relies on $\text{NAM}$ cancelling themselves to give the identity vector – and the same thing for the other two cases with $\text{CAP}$ and $\text{MON}$ cancelling themselves.
However, very few vectors in the HRR algebra are their own inverses. In fact, there are only $2^d$ such vectors. These are the vectors where each Fourier coefficient is either $-1$ or $1$ (such that each complex number either rotates itself to become the identity 1, or stays at 1, respectively).
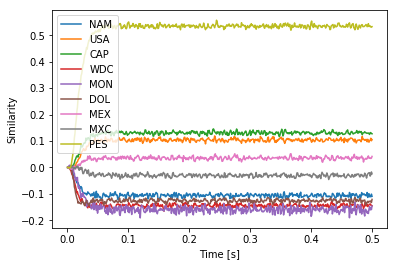
One work-around is to construct the query vector a little differently to fit the example and the HRR algebra. In particular, if we instead construct the query as DOL * MON * MON then its inverse will take care of the two MON that were bound together in $F_{UM}$. With 64 dimensions, this seems to work okay most of the time.
d = 64
with spa.Network(seed=0) as model:
# Maybe this will help?
model.config[spa.State].neurons_per_dimension = 200
fum = spa.Transcode('(NAM*USA + CAP*WDC + MON*DOL) * '
'(NAM*MEX + CAP*MXC + MON*PES)', output_vocab=d)
query = spa.Transcode('DOL * MON * MON', output_vocab=d)
result = spa.State(vocab=d)
fum * ~query >> result
p = nengo.Probe(result.output, synapse=0.01)
with nengo.Simulator(model) as sim:
sim.run(0.5)
plt.plot(sim.trange(), spa.similarity(sim.data[p], result.vocab))
plt.xlabel("Time [s]")
plt.ylabel("Similarity")
plt.legend(result.vocab, loc='best')
plt.show()
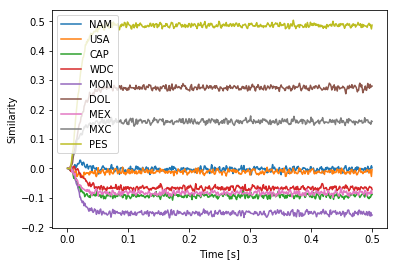
Another possibility is to pick a vector for MON that is its own inverse, i.e., satisfying MON = ~MON or equivalently MON * MON = 1, where 1 is the identity vector. Again, there are $2^d$ such vectors. To keep things simple, let’s just pick MON = -1. This also seems to work well most of the time.
vocab = spa.Vocabulary(
dimensions=64, pointer_gen=np.random.RandomState(seed=0))
# Add the vectors in the same order as before
# to get consistent colours / legend
vocab.populate('NAM; USA; CAP; WDC')
vocab.add('MON', vocab.parse('-1'))
vocab.populate('DOL; MEX; MXC; PES')
# Check that it is its own inverse
assert np.allclose(vocab.parse('MON').v,
vocab.parse('~MON').v)
# Check that it cancels out to give the identity
assert np.allclose(vocab.parse('MON * MON').v,
vocab.parse('1').v)
with spa.Network(seed=0) as model:
model.config[spa.State].neurons_per_dimension = 200
fum = spa.Transcode('(NAM*USA + CAP*WDC + MON*DOL) * '
'(NAM*MEX + CAP*MXC + MON*PES)',
output_vocab=vocab)
query = spa.Transcode('DOL', output_vocab=vocab)
result = spa.State(vocab=vocab)
fum * ~query >> result
p = nengo.Probe(result.output, synapse=0.01)
with nengo.Simulator(model) as sim:
sim.run(0.5)
plt.plot(sim.trange(), spa.similarity(sim.data[p], result.vocab))
plt.xlabel("Time [s]")
plt.ylabel("Similarity")
plt.legend(result.vocab, loc='best')
plt.show()