Hi @jone. This is a common problem in computational neuroscience, and any of the approaches that apply elsewhere can also apply to Nengo.
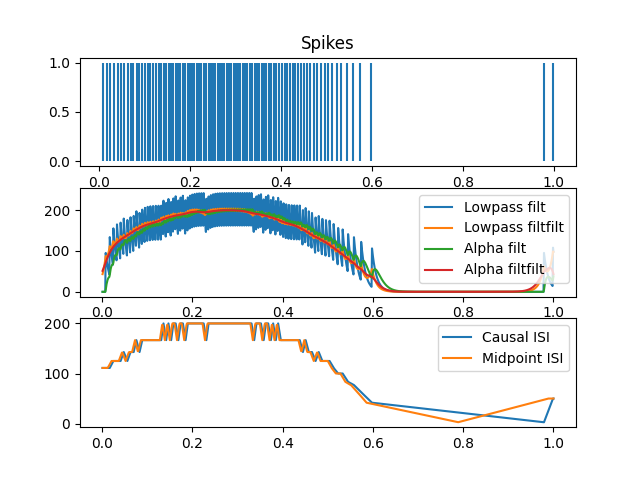
The two main approaches that come to mind for me are 1) kernel-based approaches, and 2) ISI-based approaches. I’ve put a script down the bottom demonstrating these approaches.
For kernel-based approaches, you’re basically convolving some sort of low-pass filtering kernel with the spike output. In Nengo, we typically do this with a Synapse class. Lowpass is a basic first-order low-pass filter, and Alpha is a second-order type of low-pass filter. You can apply them causally with filt, or apply them both forward and back (which is non-causal) with filtfilt; this latter approach will result in no delay. You could also apply something like a Gaussian kernel; we don’t have a Synapse for this but you could create a Gaussian kernel and convolve it with the spikes manually.
For the ISI-based approaches, you compute the inter-spike intervals (ISIs), which is the time between one spike and the next, and then use the inverse of these intervals as the firing rate. Again, you can do this in a causal way, or in a more centred way that is non-causal.
import matplotlib.pyplot as plt
import nengo
import numpy as np
with nengo.Network() as net:
u = nengo.Node(lambda t: np.sin(6 * t))
a = nengo.Ensemble(1, 1, encoders=[[1]], intercepts=[-0.5], max_rates=[200])
nengo.Connection(u, a, synapse=None)
p = nengo.Probe(a.neurons)
with nengo.Simulator(net) as sim:
sim.run(1.0)
t = sim.trange()
spikes = sim.data[p]
spike_times = t[spikes[:, 0] > 0]
lowpass_spikes = nengo.synapses.Lowpass(tau=0.01).filt(spikes)
lowpass_ff_spikes = nengo.synapses.Lowpass(tau=0.01).filtfilt(spikes)
alpha_spikes = nengo.synapses.Alpha(tau=0.01).filt(spikes)
alpha_ff_spikes = nengo.synapses.Alpha(tau=0.01).filtfilt(spikes)
isi = np.diff(spike_times)
causal_isi = np.interp(t, spike_times[1:], 1 / isi)
midpoint_isi = np.interp(t, 0.5 * (spike_times[:-1] + spike_times[1:]), 1 / isi)
plt.subplot(311)
plt.vlines(spike_times, 0, 1)
plt.title("Spikes")
plt.subplot(312)
plt.plot(t, lowpass_spikes, label="Lowpass filt")
plt.plot(t, lowpass_ff_spikes, label="Lowpass filtfilt")
plt.plot(t, alpha_spikes, label="Alpha filt")
plt.plot(t, alpha_ff_spikes, label="Alpha filtfilt")
plt.legend()
plt.subplot(313)
plt.plot(t, causal_isi, label="Causal ISI")
plt.plot(t, midpoint_isi, label="Midpoint ISI")
plt.legend()
plt.show()