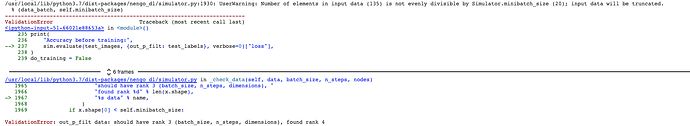
Hi, I am quite new to nengo. I was following the “Optimizing a spiking neural network” example in Nengo; seemed to work perfectly. Then I got into the task of importing my own dataset and got a lot of errors. The sample dataset which I was working with is the dogs and cats dataset which can be found at this link . I have tried numerous ways of importing my own dataset, but none of the methods seem to be working. Here is the algorithm which I’m using to replace the current method to load the dataset.
Instead of this:
(train_images, train_labels), (
test_images,
test_labels,
) = tf.keras.datasets.mnist.load_data()
I was doing this to load a dataset:
plt.figure(figsize=(28,28))
test_folder=r'/data/train/cats'
IMG_WIDTH=28
IMG_HEIGHT=28
img_folder=r'/data/train'
for i in range(5):
file = random.choice(os.listdir(test_folder))
image_path= os.path.join(test_folder, file)
img=mpimg.imread(image_path)
ax=plt.subplot(1,5,i+1)
ax.title.set_text(file)
plt.imshow(img)
### Shuffle Dataset and labels
def unison_shuffled_copies_train(a, b):
assert len(a) == len(b)
p = np.random.permutation(len(a))
return a[p], b[p]
def unison_shuffled_copies_test(a, b):
assert len(a) == len(b)
p = np.random.permutation(len(a))
return a[p], b[p]
def create_dataset_train(img_folder):
img_data_array=[]
class_name=[]
for dir1 in os.listdir(img_folder):
for file in os.listdir(os.path.join(img_folder, dir1)):
image_path= os.path.join(img_folder, dir1, file)
image= cv2.imread( image_path, 0)
if image is not None:
image=cv2.resize(image, (IMG_HEIGHT, IMG_WIDTH),interpolation = cv2.INTER_AREA)
image=np.array(image)
image = image.astype('float32')
image /= 255
img_data_array.append(image)
class_name.append(dir1)
return img_data_array, class_name
# extract the image array and class name
img_data, class_name =create_dataset_train(r'/data/train')
train_images = np.array(img_data)
print("Train I: ", train_images.shape)
target_dict={k: v for v, k in enumerate(np.unique(class_name))}
target_val= [target_dict[class_name[i]] for i in range(len(class_name))]
train_labels = np.array(target_val)
print(train_labels)
train_images, train_labels = unison_shuffled_copies_train(train_images,train_labels)
img_folders = "/data/test"
def create_dataset_test(img_folders):
img_data_array=[]
class_name=[]
for dir2 in os.listdir(img_folders):
for file in os.listdir(os.path.join(img_folder, dir2)):
image_path= os.path.join(img_folder, dir2, file)
image= cv2.imread( image_path, 0)
if image is not None:
image=cv2.resize(image, (IMG_HEIGHT, IMG_WIDTH),interpolation = cv2.INTER_AREA)
image=np.array(image)
image = image.astype('float32')
image /= 255
img_data_array.append(image)
class_name.append(dir2)
return img_data_array, class_name
# extract the image array and class name
img_data, class_name =create_dataset_test(r'/data/test')
for i in range(25):
plt.subplot(5, 5, i + 1)
plt.imshow(train_images[i].reshape(28,28), cmap=plt.cm.binary)
plt.axis("off")
test_images = np.array(img_data)
target_dict={k: v for v, k in enumerate(np.unique(class_name))}
target_val= [target_dict[class_name[i]] for i in range(len(class_name))]
test_labels = np.array(target_val)
test_images, test_labels = unison_shuffled_copies_test(test_images,test_labels)
train_images = train_images.reshape((train_images.shape[0], -1))
test_images = test_images.reshape((test_images.shape[0], -1))
plt.figure(figsize=(12, 4))
for i in range(3):
plt.subplot(1, 3, i + 1)
plt.imshow(np.reshape(train_images[i], (28, 28)), cmap="gray")
plt.axis("off")
plt.title(str(train_labels[i]))
And then from there on the rest of the code is run, which is the same code from the example MNIST one. I am not entirely sure if this is the correct way of importing a dataset for this Nengo-specific model; whether I am just over-complicating it. I want it to be in the exact format as the MNIST dataset, which I checked and in the end it looks like its the same format but there’s still errors in shape e.t.c. Could anyone help me please? Is there any pre-written piece of code that is good to load these type of datasets. As a note, I am also loading in the images as grayscale for simplicity. If there’s anything unclear with what I defined in the problem, please let me know and I can edit this.Thanks in advance.