I am tring to implement a regression problem which I will eventually implement on Loihi. I started with the MNIST classification problem example and tried to modify the compile section.
I replaced the RMSprop(0.001), SparseCategoricalCrossentropy, and sparse_categorical_accuracy with
Adam(0.001), MeanSquaredError, and Accuracy (as I used in Neural network model)
Unfortunately I am getting 0% accuracy unless I convert it as classification problem
Input: 4 x 3 and Output: 1 x 3
My code is attached below:
import warnings
import matplotlib.pyplot as plt
import nengo
import nengo_dl
import nengo_loihi
import numpy as np
import tensorflow as tf
from tensorflow.keras.models import Sequential, clone_model
from tensorflow.keras.layers import Input,Dense, Dropout, Activation, Flatten,Conv2D
from tensorflow.keras.layers import MaxPooling2D
from tensorflow.keras.datasets import mnist
from tensorflow.keras.utils import normalize
from urllib.request import urlretrieve
import pickle
# ignore NengoDL warning about no GPU
warnings.filterwarnings("ignore", message="No GPU", module="nengo_dl")
np.random.seed(0)
tf.random.set_seed(0)
num_classes = 3
# load dataset
train_X= np.array([[[ 0., 0., 1.],
[ 0., 1., 1.],
[ 0., 2., 1.],
[ 1., -1., 0.]],
[[ 0., 0., 1.],
[ 0., 1., 1.],
[ 0., 2., 1.],
[ 2., -1., 0.]],
[[ 0., 0., 1.],
[ 0., 1., 1.],
[ 0., 2., 1.],
[ 3., -1., 0.]]])
train_Y = np.array([[0.029, 0.059, 0.079],
[0.298, 0.985, 0.546],
[0.854, 0.911, 0.405]])
# Label modification like Mnist
labels = []
for i in range(train_Y.shape[0]):
output = np.argmax(train_Y[i])
labels.append(int(output))
labels = np.array(labels)
train_images_R = train_X.reshape((train_X.shape[0],train_X.shape[1]*train_X.shape[2])) #NO need
train_labels_R = train_Y.copy() #NO need
train_Images = train_X.reshape((train_X.shape[0], 1, -1)) # 3,4,3 => 25,1,12
train_Labels = train_Y.reshape((train_Y.shape[0], 1, -1)) # 3,3 => 3,1,3
train_Labels_RM = labels.reshape((labels.shape[0], 1, -1)) # 3, => 3,1,1
def modelDef():
inp = tf.keras.Input(shape=(4, 3, 1), name="input")
# transform input signal to spikes using trainable 1x1 convolutional layer
to_spikes_layer = tf.keras.layers.Conv2D(
filters=3, # 3 neurons per pixel
kernel_size=1,
strides=1,
activation=tf.nn.relu,
use_bias=False,
name="to-spikes",
)
to_spikes = to_spikes_layer(inp)
# on-chip layers
flatten = tf.keras.layers.Flatten(name="flatten")(to_spikes)
dense0_layer = tf.keras.layers.Dense(units=10, activation=tf.nn.relu, name="dense0")
dense0 = dense0_layer(flatten)
# since this final output layer has no activation function,
# it will be converted to a `nengo.Node` and run off-chip
dense1 = tf.keras.layers.Dense(units=num_classes, name="dense1")(dense0)
model = tf.keras.Model(inputs=inp, outputs=dense1)
model.summary()
return model
def train(params_file="./keras_to_loihi_params123", epochs=1, **kwargs):
model = modelDef()
miniBatch = 3
converter = nengo_dl.Converter(model, **kwargs)
with nengo_dl.Simulator(converter.net, seed=0, minibatch_size=miniBatch) as sim:
'''
Followed MNIST classification structure
'''
# sim.compile(
# optimizer=tf.optimizers.RMSprop(0.001),
# loss= {
# converter.outputs[model.get_layer('dense1')]: tf.losses.SparseCategoricalCrossentropy(
# from_logits=True
# )
# },
# metrics={converter.outputs[model.get_layer('dense1')]: tf.metrics.sparse_categorical_accuracy},
# )
# sim.fit(
# {converter.inputs[model.get_layer('input')]: train_Images},
# {converter.outputs[model.get_layer('dense1')]: train_Labels_RM},
# epochs=epochs,
# )
'''
Not followed MNIST structure
'''
sim.compile(
optimizer=tf.optimizers.Adam(0.001),
loss= {
converter.outputs[model.get_layer('dense1')]: tf.losses.MeanSquaredError()
},
metrics={converter.outputs[model.get_layer('dense1')]: tf.metrics.Accuracy()},
)
sim.fit(
{converter.inputs[model.get_layer('input')]: train_Images},
{converter.outputs[model.get_layer('dense1')]: train_Labels},
epochs=epochs,
)
# train this network with normal ReLU neurons
train(
epochs=2000,
swap_activations={tf.nn.relu: nengo.RectifiedLinear()},
)
Could you please suggest me how to fix the issue (compile section)?
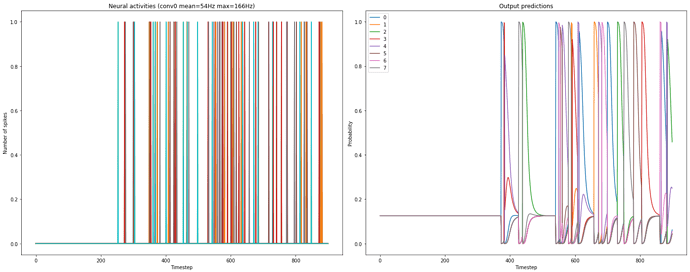
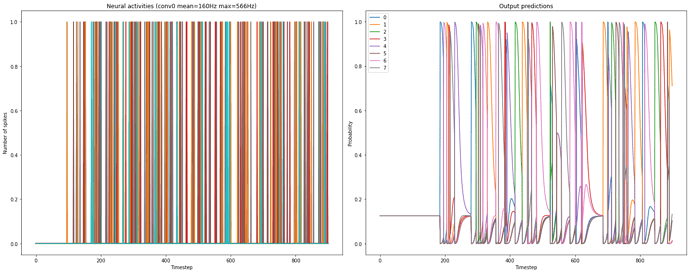
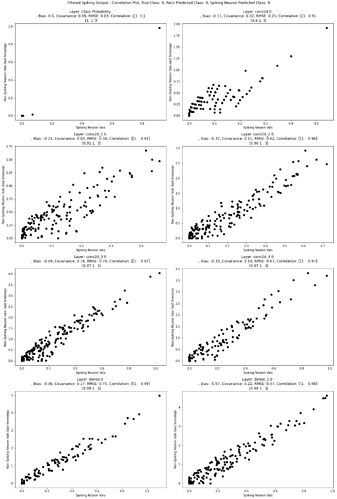
 ). With respect to plotting the spikes, I would suggest you to go step by step, and try to understand how spikes are calculated/represented.
). With respect to plotting the spikes, I would suggest you to go step by step, and try to understand how spikes are calculated/represented.