Hi @HowardC, and welcome back!
For future reference, the Nengo forums allows you to edit your posts! ![]()
You can edit posts by clicking on the “…” button right next to “Reply”, and then clicking on the pencil button. Deleting a post is done similarly, by pressing the “…” button, then the garbage can button.
Now, let’s get to answering your questions!
This is a fantastic conceptual question to ask, and the quick answer is no. If you have a recurrent (decoded) connection, every neuron in ens_A connects to every neuron in ens_A, including itself. To see why this is the case, let us consider the more straight-forward 2-ensemble connection. Consider the following Nengo code:
ens_A = nengo.Ensemble(5, 1)
ens_B = nengo.Ensemble(5, 1)
nengo.Connection(ens_A, ens_B)
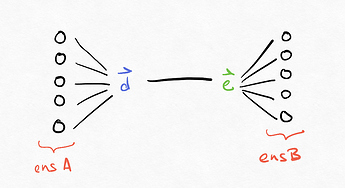
Conceptually, what this amounts to is something like this (please excuse my crude handdrawn diagrams), where \vec{d} are the decoders for ens_A and \vec{e} are the encoders for ens_B.
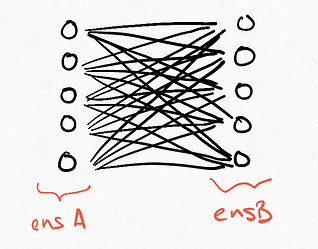
Mathematically, the encoders and decoders can be combined to form a fully connected weight matrix, so the diagram below is the mathematical equivalent of the network:
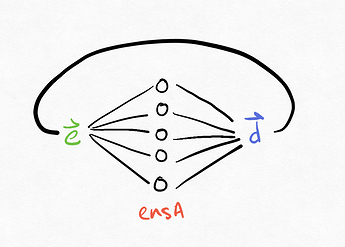
Now, to understand what is happening in the case of a recurrent connection, we can consider a recurrent connection as a rolled up version of a 2-ensemble connection, where the neurons in ens_A and ens_B are the same. The decoded version of a recurrent connection the looks like this:
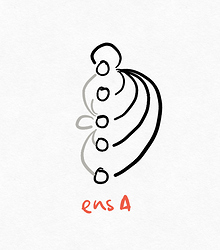
Converting this into the fully connected weight matrix version yields this (I’ve only drawn the connections for the two neurons, it gets messy if I include all of the connections. I’ve coloured the connections differently to differentiate the source neuron):