I don’t understand the way how to calculate the accuracy of the model and predict the output. The two below models have the same way that calculates the accuracy and predicts output value but one model works and the other doesn’t. Can anybody tell me the reason for this, please
Ensemble_SNN.ipynb (21.5 KB)
Ensemble_SNN_1.ipynb (32.2 KB)
?
Hi @ntquyen2,
Comparing the two notebooks, it seems like the networks are different between the two cases. For example, the Nengo network in the Ensemble_SNN notebook does not have the convolution transforms between each layer like the Ensemble_SNN1 notebook does.
Nengo does have a built-in convolution transform as well, and an example of it’s use can be found in the MNIST convolution network example for NengoLoihi. Note that while the NengoLoihi example does target the Loihi platform, the syntax for the convolution transform is the same across the various Nengo platforms.
Also absent are the average pooling transforms. In Nengo, you’d implement this using the transform parameter on the nengo.Connection objects.
As a side note, if you are trying to reimplement your CNN as a spiking neural network, I’d advise you check out our NengoDL example on how to use the NengoDL converter to convert a CNN to an equivalent SNN.
Hi @xchoo
I am grateful to you and thanks for your reply. Also, I don’t understand what you said " Also absent are the average pooling transforms. In Nengo, you’d implement this using the transform parameter on the nengo.Connection objects." because I think nengo_dl. Layer that integrates nengo.TensorNode and nengo.Connection as Coming from TensorFlow to NengoDL — NengoDL 3.4.3.dev0 docs at Inserting TensorFlow code. In addition, in the Ensemble_SNN1 notebook, I want to plot output values as an example Deep learning parameter optimization on the above link but I don’t how to plot. I coded as that example but the error occurs as you see in the notebook
Hi @xchoo
Can you help me please?
Hi @xchoo
Sorry If I bother you. I saw the example MNIST convolutional network — NengoLoihi 1.1.0.dev0 docs, however I wonder that when increase firing rate, whether the trade off happens or not. For instance, accuracy or the activities of neurons
Hi @xchoo
Sorry you due to this inconvenience. I run my code as LOIHI example and add skip connection in my network, my network run however when I simulate one, the error occurs ValueError: A Concatenate layer requires inputs with matching shapes except for the concat axis. Got inputs shapes: [(200, 6272), (0, 6272)]. I think when the network is OK, everything does. Can you tell me the reason for that, please? Thank you very much
MyEnsemble_Loihi.ipynb (50.9 KB)
My comment about the average pooling transforms is in regard to the comparison between the two notebooks you provided. You are correct in the understanding that if you do something like this:
x = nengo_dl.Layer(tf.keras.layers.AveragePooling2D(pool_size=2, strides=2))(
x,shape_in=(14,14,64))
your Nengo network will contain an average pooling layer. However, if you look at the network (net) in the Ensemble_SNN notebook, you’ll see that it only contains an input node, and two nengo.Ensemble layers (which are equivalent to the nengo_dl.Layer(neuron_type)(x) layers in the Ensemble_SNN1 notebook). Thus, the Ensemble_SNN network is missing the convolution layers, and the average pooling layers, which is probably why one network works, while the other doesn’t. If you want both notebooks to have the same network structure, you’ll need to implement both the convolution transforms (see the example notebook I posted in my previous reply), and the average pooling transform.
I’m not entirely sure what tradeoff you are asking about here, so I’m going to answer to the best of my understanding of your question. For spiking neural networks, part of the drop in accuracy you see (when compared to “conventional” neural networks) is due to the extra noise the spiking behaviour adds to the network. The noise is due to the fact that spiking neurons cannot represent a constant value. Rather, they can only produce values a certain frequency (whenever they produce a spike), and at all times when they are not spiking, their outputs are zero. To increase the accuracy of the network, one can introduce smoothing to the spike trains. Biologically, this is the post-synaptic current applied at synapses between each neuron. In Nengo, the smoothing is accomplished by using the synapse parameter on any nengo.Connection.
Alternatively, spike noise can be reduced by increasing the firing rate of the neurons. If your neurons can fire infinitely fast (or as fast as 1 spike per simulation timestep), they essentially become rate neurons. However, increasing spiking rates decreases the advantages of having spiking neurons in the first place, which is the reduction in energy usage (particularly on a chip like Loihi). Our Keras-to-SNN NengoDL example walks through these concepts using an example network.
I believe that this is because you have not specified the slicing of the x tensor properly. The first dimension of the tensor should be the minibatch size, so what you want to be slicing is the second dimension, like so:
x0 = x[:, :layer4.size_out]
x1 = x[:, layer4.size_out:]
However, once you have fixed that issue, there seems to be other issues with your network as well. From a quick glance, the layer input and output sizes don’t seem to match up properly, so you’ll need to fix that before the network can be run. This post is using a similar function (the Tensorflow add layer), and may help you.
I am really really glad to your appreciation so much @xchoo. And I wonder that whether Nengo can use Modelcheckpoint to save best weight or not because I use EarlyStopping and it works but ModelCheckpoint doesn’t when training.
Ensemble_SNN.ipynb (127.8 KB)
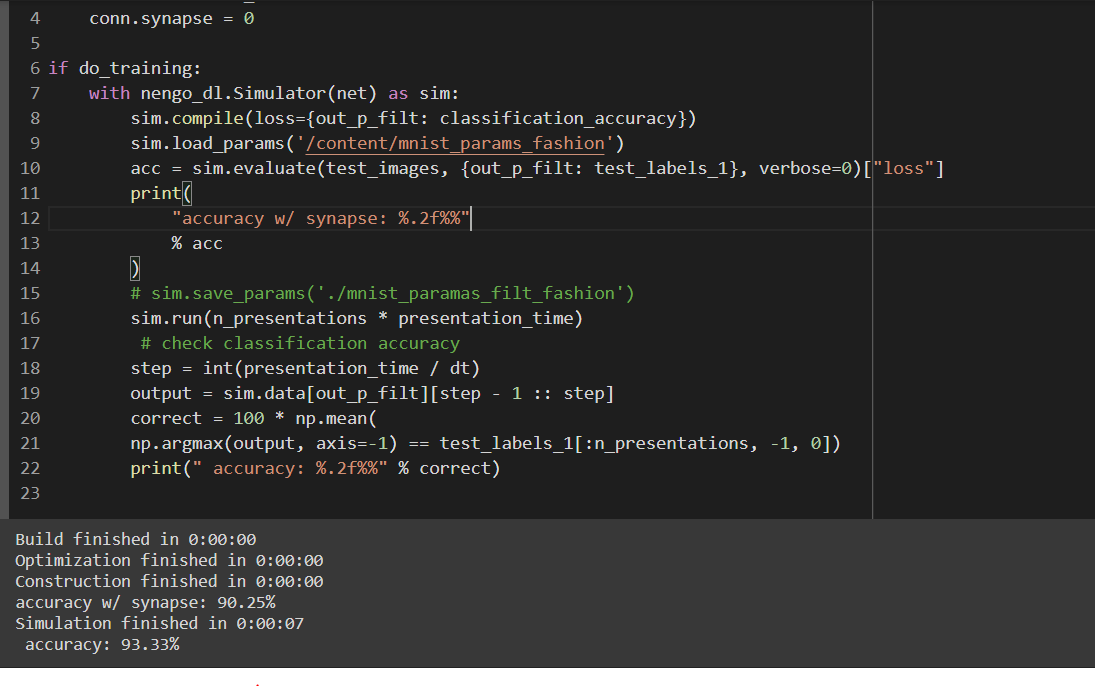
Sorry, @xchoo if I bother you. However, I wonder that when I use sim.evaluate and sim.data to evaluate on atest images, both give different results so what gives the right results? I see the example MNIST Loihi, they use both of the approaches to evaluate the test image. The notebook is attached below. Can you explain me, please? Maybe when I use sim.data with the number of test images (not all test_labels_1) so the accuracy can increase?
Ensemble_SNN.ipynb (49.8 KB)
Both should reflect what is happening with the network. In essence, both are “correct”. Looking at your code, I can see that both the sim.evaluate function, and the plot from sim.data are logically consistent. Let me explain what I think is happening:
You modified the test labels, and the network to output only 2 classes: “0” for all images that are the image of a zero, and “1” for all other digits. After training the sim.evaluate function returns an accuracy of 93%. If you look at the sim.data plot, however, you see that all the network is doing is outputting 1 for pretty much all of the test images. So, you might be wondering, how can the network achieve an overall accuracy of 93%, but yet the sim.data plot look so wrong (it seems to be consistently wrong with the “zero” images)?
That comes down to how the data is structured. Since you are using the MNIST dataset, there is an equal (or roughly equal) number of 0’s, 1’s, 2’s, etc. You also reconfigured the network and training to classify all digits that are not 0 with the label “1”. Now, consider what happens if your network just outputs “1” all of the time. For the modified data labels, the label “1” is correct for 90% of all of the images since 90% of all the images are not-a-zero. I think this is what is happening here.
If you want the network to be better at distinguishing between zeros and non-zeros, I think you need to modify the training data such that there are an equal number of zeroes and non-zeros.