(See next post for quicker to answer questions that don’t need this context)
Dear all,
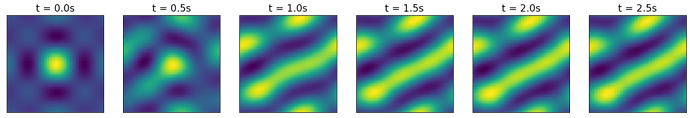
I am implementing a head direction network in Nengo, based on this publication: http://compneuro.uwaterloo.ca/files/publications/conklin.2005.pdf. In short, the neurons represent the first n components of the Fourier Series of a set of Gaussians, where each Gaussian’s center represents a different head direction on a cyclic axis (or in the paper a different location on a toroidal grid). The currently represented Gaussian can be shifted around by a rotation matrix applied by recurrent connections.
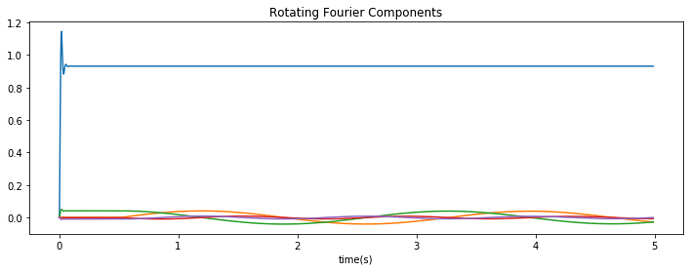
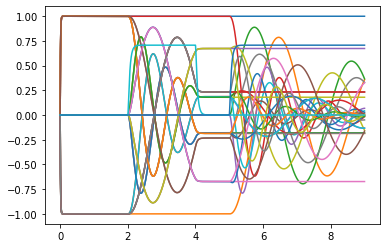
I managed to get everything to work using Direct neurons; the representation of the network, when a constant rotation speed is applied, looks like this:
The DC component in blue remains constant, while the other components oscillate as expected.
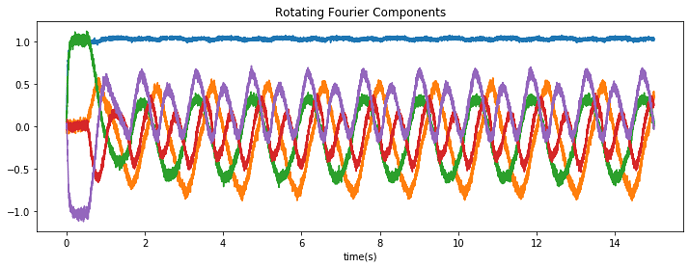
I am however struggling to get the network to work using LIF neurons, there are two problems I run into. The first is that when I set the encoders to the Fourier components of the Gaussians as in the paper (where each neuron represents a Gaussian with a different mean), the resulting activation when a Gaussian is provided as input is not a Gaussian anymore. My guess is that this is due to the scaling/normalization that is applied to the encoders, should I take this into account somehow?
Second, the authors of the paper managed to get a stable recurrent activity with a rather small time constant (0.005), which is not happening at all in my network; I only get some stable activity if increase the time constant to way higher. Is this just due to the bad representation of my neurons (I guess a longer time constant smooths out imperfections more)?
The code can be found here: https://github.com/Matthijspals/NengoSLAM/blob/master/HD_network.ipynb
Thank you so much for taking the time to look at this and please let me know if anything is unclear!