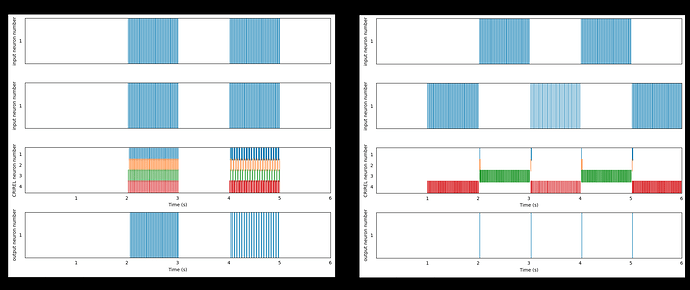
I have built a very simple recurrent circuit in NengoLoihi. I have two very simple piecewise inputs into the network. There is no noise in my model . However each time I run the code, the output of the network is different each time. I change nothing between each run, yet the output does. My network I am building is highly recurrent, and should show hysteresis. However, I would expect that the initial conditions of LOIHI would be the same every time I run my script. This doesn’t seem to be the case. Is there some line of code that makes sure the initial conditions are the same each time?
Below I have attached the code and the two different outputs that it flips between.
import matplotlib.pyplot as plt
import nengo
import nengo_loihi
import numpy as np
from nengo.processes import Piecewise
from nengo.utils.matplotlib import rasterplot
nengo_loihi.set_defaults()
with nengo.Network() as model:
# Logic gate neurons#
stim1 = nengo.Node(
Piecewise({0: 0, 1: 1, 2: -1, 3: 1, 4: -1, 5: 0.5, 6: -1}), size_out=1
)
stim2 = nengo.Node(
Piecewise({0.1: 0, 1: 1, 2: -1, 3: 0.5, 4: -1, 5: 1, 6: -1}), size_out=1
)
input1 = nengo.Ensemble(
1,
dimensions=1,
neuron_type=nengo_loihi.neurons.LoihiLIF(tau_rc=0.02),
gain=[1],
bias=[1],
)
input2 = nengo.Ensemble(
1,
dimensions=1,
neuron_type=nengo_loihi.neurons.LoihiLIF(tau_rc=0.02),
gain=[1],
bias=[1],
)
CRIREL = nengo.Ensemble(
4,
dimensions=1,
neuron_type=nengo_loihi.neurons.LoihiLIF(tau_rc=0.02),
gain=np.ones(4),
bias=[0.1, 0.1, 0.5, 0.5],
)
output = nengo.Ensemble(
1,
dimensions=1,
neuron_type=nengo_loihi.neurons.LoihiLIF(tau_rc=0.02),
gain=[1],
bias=[-2],
)
nengo.Connection(stim1, input1)
nengo.Connection(stim2, input2)
tau = 0.005
inputSyn1 = 0.1 * np.array([[1], [0], [1], [0]])
inputSyn2 = 0.1 * np.array([[0], [1], [0], [1]])
nengo.Connection(input1.neurons, CRIREL.neurons, transform=inputSyn1, synapse=tau)
nengo.Connection(input2.neurons, CRIREL.neurons, transform=inputSyn2, synapse=tau)
ampa = 0.005
gaba = 0.005
gee = 0.15
gei = 0.15
gie = -0.30
gii = -0.30
CRIREL_ampa_syn = np.array(
[[0, gee, gie, 0], [gee, 0, 0, gie], [0, 0, 0, 0], [0, 0, 0, 0]]
)
CRIREL_gaba_syn = np.array(
[[0, 0, 0, 0], [0, 0, 0, 0], [gei, 0, 0, gii], [0, gei, gii, 0]]
)
nengo.Connection(
CRIREL.neurons, CRIREL.neurons, transform=CRIREL_ampa_syn, synapse=ampa
)
nengo.Connection(
CRIREL.neurons, CRIREL.neurons, transform=CRIREL_gaba_syn, synapse=gaba
)
gout = 0.025
out_syn = np.array([[gout, gout, 0, 0]])
nengo.Connection(CRIREL.neurons, output.neurons, transform=out_syn, synapse=ampa)
input1_probe = nengo.Probe(input1.neurons)
input2_probe = nengo.Probe(input2.neurons)
CRIREL_probe = nengo.Probe(CRIREL.neurons)
output_probe = nengo.Probe(output.neurons)
with nengo_loihi.Simulator(model) as sim:
sim.run(6)
t = sim.trange()
def plot_rasters(t, data):
plt.figure(figsize=(10, 8))
plt.subplot(4, 1, 1)
rasterplot(t, data[input1_probe])
plt.xticks(())
plt.ylabel("input neuron number")
plt.subplot(4, 1, 2)
rasterplot(t, data[input2_probe])
plt.xticks(())
plt.ylabel("input neuron number")
plt.subplot(4, 1, 3)
rasterplot(t, data[CRIREL_probe])
plt.ylabel("CRIREL neuron number")
plt.xlabel("Time (s)")
plt.subplot(4, 1, 4)
rasterplot(t, data[output_probe])
plt.ylabel("output neuron number")
plt.xlabel("Time (s)")
plt.tight_layout()
plot_rasters(t, sim.data)
plt.show()