Hi @FilipD
I may have a solution for you, but first, I’ll get to your questions:
That is correct. With the BCM rule, even zero-weight connections can be changed by the weight update rule.
I’m not too sure about what the performance impact would be here, but I think there will be some performance decrease since (for regular Nengo) the numpy matrix operations are not being taken advantage of with multiple connections.
While this would have slightly better performance than solution 2, it should still perform worse that solution 1 since multiple connection updates is slower than 1 numpy matrix update for one connection.
Proposed Solution
So, having thought about it for a while, I believe that I may have a solution that fits your needs. My idea is to create what is essentially a copy of the BCM rule, with one critical change. The code that does the weights update for the BCM rule is this:
def step_simbcmmasked():
delta[...] = np.outer(
alpha * post_filtered * (post_filtered - theta), pre_filtered
)
The computed delta matrix determines how the connection weight is changed (i.e., new_weight = old_weight + delta. So, for any connection that we don’t want updated, all we need to do is to zero out that corresponding value in the delta matrix. For everything else that we want updated, we just leave the value in the delta matrix there.
So, my idea is to create a separate mask matrix, and use the numpy multiply function to apply the mask to the delta matrix. If we have a 1 in the mask, the delta value is untouched, but, crucially, if we put a 0 in the mask, the corresponding value in the delta matrix is zeroed out, and thus the weight update for that connection will not happen. In essence, this code should do it:
def step_simbcmmasked():
delta[...] = np.multiply(
np.outer(alpha * post_filtered * (post_filtered - theta), pre_filtered),
mask,
)
Here is some code demonstrating the modified (masked) BCM rule, along with some test code:
- The custom BCM rule: bcm_mask.py (5.2 KB)
- The test code: test_bcm_mask.py (5.9 KB)
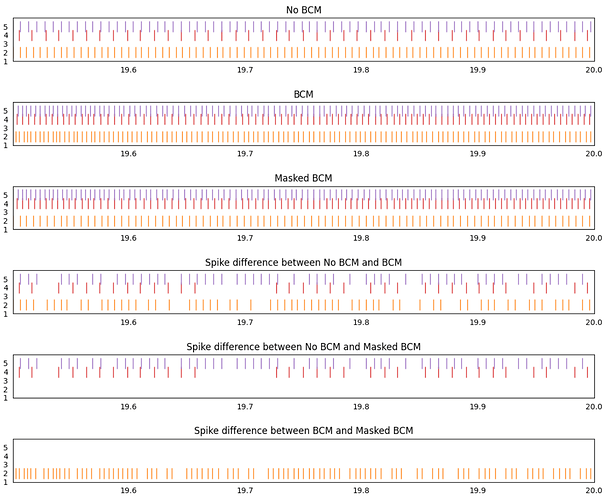
Here’s a plot demonstrating the code working:
The first 3 plots show the spiking output of a network with no learning rule, a network with the BCM learning rule, and a network with the masked BCM learning rule (where all inputs to neuron 2 have been masked).
The next 3 plots show any spiking activity difference between the different spike rasters. Any spike that appears at the same place in both spike raster will be removed from the final plot, but any spike that shows up in only one of the spike raster will show up in the final plot.
Comparing the no-bcm and bcm networks, we see that spike differences appear for all 3 active neurons in the post population. This is to be expected.
Comparing the no-bcm and masked bcm networks, we see that only neuron 4 and 5 show spike differences. Once again, this is to be expected since the weight updates for all connections into neuron 2 have been disabled, thus, the spike output of neuron 2 should be the same as in the no-bcm case.
Comparing the bcm and masked bcm networks, we see that since the bcm rule is applied to all neurons apart from neuron 2, only neuron 2 show a difference between these two cases. Since all 3 networks have the same seed, the BCM rule affects neuron 4 and 5 identically in both the bcm and masked bcm networks.
Additional Questions
The code I posted above currently only works in the Python Nengo. For NengoOCL, a custom ocl kernel is needed to implement the modified weight update. I’m not very familiar with OCL programming, so this may take some time for me to get working, although a quick look seem to indicate that it shouldn’t be too difficult. The existing BCM OCL kernel can be found here if you want to take a stab at it yourself. When I get the chance to test the OCL version I’ll update this thread.
Hmmm. Not really. Unlike something like tensorflow, Nengo doesn’t have a concept of “physical” arrangements of neurons within an ensemble. That being said, the advantage of that is that one ensemble can be physically interpreted as being any physical dimensionality (even a 3D arrangement if you want). But, the downside is that the physical arrangement of the neurons is up to the user to determine, by using the appropriate transformation matrices between connections. I’ll consult the other Nengo devs to see if they have any other ideas. ![]()