Hi!
My project consists of translating a biologically detailed model of Basal Ganglia from NEST to Nengo. It is defined in terms of biological data (e.g. average number of connections from population A to population B, capacitance of A’s neurons, postsynaptic potentials…), not in terms of mathematical expressions. Therefore, I cannot really use NEF’s encoders and decoders. In this model, a command is represented by the activity of a discrete channel of neurons in ganglia. Once I implemented the model in Nengo using Direct Connections, will I be able to use it functionally (i.e. having inputs and outputs encodable and decodable by NEF)?
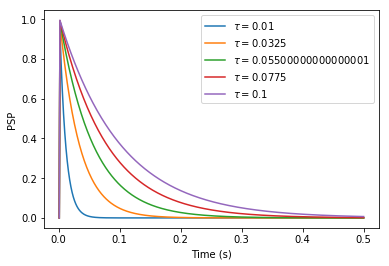
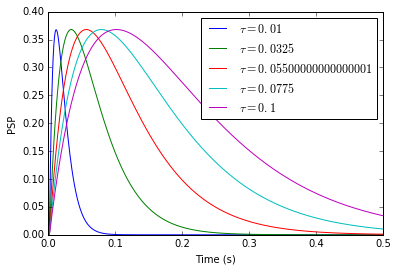
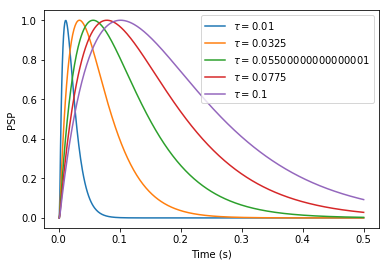
Moreover, I do not really understand how to choose the postsynaptic potential of a Direct Connection. In NEST, the following code creates a connection between population A and B that will cause postsynaptic potentials (PSP) of 1 mV in neurons of B. Amplitude of PSPs is therefore independent of synaptic time constants.
nest.SetStatus(A, {'tau_syn':[5.]}) # set synaptic time constant of receptor type 1
nest.Connect(A, B, syn_spec={'receptor_type':1, 'weight':1}) # connect A->B with weight (PSP) = 1
However, when I try to connect neurons of two populations in Nengo, the input of B seems to depend on the time constant of the synapse. I would expect the following code to exhibit the same behavior as NEST, with weights specified in the transform parameter:
syn_AMPA = nengo.synapses.Alpha(tau_syn_AMPA/1000.)
nengo.Connection(A.neurons, B.neurons, transform=[1], synapse=syn_AMPA)
But when I probe the input of B, this is what I get:
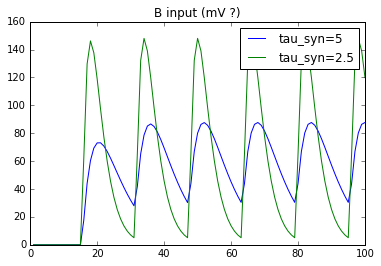
NB: transform does scale the input. But I would like to use it as an absolute value of PSP.
Can you please tell me how to properly set the weight/PSP of a direct connection and whether or not I will be able to encode/decode values?