Hi!
I was trying to create a fully connected layer from neurons of one ensemble (say X with 10 neurons) to neurons of other (say Y, also having 10 neurons), and I tried using nengo.Connection(X.neurons, Y.neurons, transform = weights), since I already have the weights (10*10 matrix).
I got an error with the dimension of weights, and I believe this is because the Connection(X.neurons, Y.neurons) makes a One-One connection, and hence the weights were expected to be of dimension [10,1]. I wanted to know how I can create an All-to-All connection of X.neurons to Y.neurons, using the weight matrix. I wanted to do this with Nengo and not NengoDL.
Hi @RohanAj!
Using .neurons in a connection is the correct way to create a fully connected layer in Nengo. As an example, the following code creates two 10-neuron ensembles, and creates a full connection weight matrix of 1’s between the two ensembles:
import nengo
import numpy as np
with nengo.Network() as model:
ens1 = nengo.Ensemble(10, 1)
ens2 = nengo.Ensemble(10, 1)
nengo.Connection(ens1.neurons, ens2.neurons, transform=np.ones((10, 10)))
with nengo.Simulator(model) as sim:
sim.run(1)
If you can provide a sample of the code while is failing, I can take a look at it to see what the issue with it is. 
Hi @xchoo,
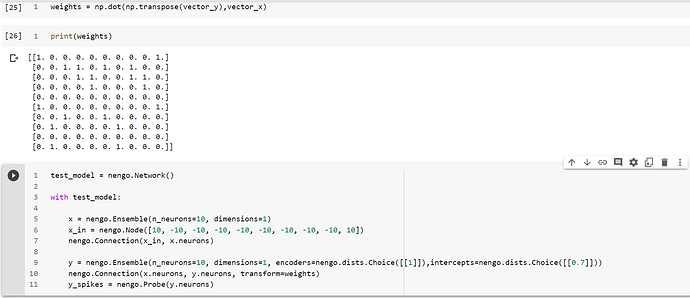
PFA the code:
weights = [[1. 0. 0. 0. 0. 0. 0. 0. 0. 1.]
[0. 0. 1. 1. 0. 1. 0. 1. 0. 0.]
[0. 0. 0. 1. 1. 0. 0. 1. 1. 0.]
[0. 0. 0. 0. 1. 0. 0. 0. 1. 0.]
[0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]
[1. 0. 0. 0. 0. 0. 0. 0. 0. 1.]
[0. 0. 1. 0. 0. 1. 0. 0. 0. 0.]
[0. 1. 0. 0. 0. 0. 1. 0. 0. 0.]
[0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]
[0. 1. 0. 0. 0. 0. 1. 0. 0. 0.]]
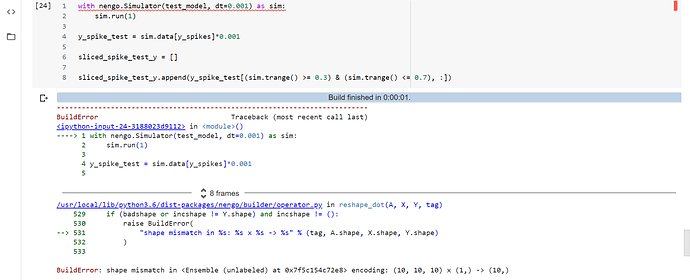
test_model = nengo.Network()
with test_model:
x = nengo.Ensemble(n_neurons=10, dimensions=1)
x_in = nengo.Node([10, -10, -10, -10, -10, -10, -10, -10, -10, 10])
nengo.Connection(x_in, x.neurons)
y = nengo.Ensemble(n_neurons=10, dimensions=1, encoders=nengo.dists.Choice([[1]]),intercepts=nengo.dists.Choice([[0.7]]))
nengo.Connection(x.neurons, y.neurons, transform=weights)
y_spikes = nengo.Probe(y.neurons)
with nengo.Simulator(test_model, dt=0.001) as sim:
sim.run(1)
y_spike_test = sim.data[y_spikes]*0.001
sliced_spike_test_y = []
sliced_spike_test_y.append(y_spike_test[(sim.trange() >= 0.3) & (sim.trange() <= 0.7), :])The error in your code seems to be in the creation of the y ensemble, specifically when you specify the intercepts. If you change your code from:
intercepts=nengo.dists.Choice([[0.7]])
to
intercepts=nengo.dists.Choice([0.7])
your code should then compile.
The Choice distributions expects a list of values from which to choose from. Additionally, the intercepts parameter expects a scalar value to be provided for each neuron. Thus, if you want to provide a Choice distribution to the intercepts parameter, it should be formatted as a list of scalar values (i.e., Choice([0.7])). Using Choice([[0.7]]) provides it a list (with one item) of 1D arrays which the builder has a hard time interpreting as a valid value for the intercept parameter (hence the error).
As a side note, when doing neuron-to-neuron connections, the encoders for the post population (i.e., the population that is the destination for the connection) is ignored, so you don’t need to specify the encoders parameter in the creation of the y ensemble.
Just an afterthought… I admit the builder error is a little dense to parse (it has a lot of additional information that clutters the output), but here’s where the indication of the error being an ensemble creation error rather than a connection error is. The full traceback was:
Traceback (most recent call last):
File "test_full_connect2.py", line 26, in <module>
with nengo.Simulator(test_model, dt=0.001) as sim:
File "/mnt/d/Users/xchoo/GitHub/nengo/nengo/simulator.py", line 169, in __init__
self.model.build(network, progress=pt.next_stage("Building", "Build"))
File "/mnt/d/Users/xchoo/GitHub/nengo/nengo/builder/builder.py", line 134, in build
built = self.builder.build(self, obj, *args, **kwargs)
File "/mnt/d/Users/xchoo/GitHub/nengo/nengo/builder/builder.py", line 239, in build
return cls.builders[obj_cls](model, obj, *args, **kwargs)
File "/mnt/d/Users/xchoo/GitHub/nengo/nengo/builder/network.py", line 78, in build_network
model.build(obj)
File "/mnt/d/Users/xchoo/GitHub/nengo/nengo/builder/builder.py", line 134, in build
built = self.builder.build(self, obj, *args, **kwargs)
File "/mnt/d/Users/xchoo/GitHub/nengo/nengo/builder/builder.py", line 239, in build
return cls.builders[obj_cls](model, obj, *args, **kwargs)
File "/mnt/d/Users/xchoo/GitHub/nengo/nengo/builder/ensemble.py", line 244, in build_ensemble
DotInc(
File "/mnt/d/Users/xchoo/GitHub/nengo/nengo/builder/operator.py", line 594, in __init__
self.reshape = reshape_dot(
File "/mnt/d/Users/xchoo/GitHub/nengo/nengo/builder/operator.py", line 538, in reshape_dot
raise BuildError(
nengo.exceptions.BuildError: shape mismatch in <Ensemble (unlabeled) at 0x7fc3ee75f190> encoding: (10, 10, 10) x (1,) -> (10,)
And if you look 3 steps before the end of the traceback, you’ll see the call throwing the error was build_ensemble. If it had been a connection misconfiguration, you would have seen an error being thrown by build_connection instead. 
Hi @xchoo, thanks a lot, it’s working now!
Also, you mentioned that I do not need to specify the encoders. However, the tuning curves change with the encoders, so won’t that be a problem?
Right, the tuning curve plot takes the encoders into account. However, since you are using only neuron-to-neuron connections in your code, you’ll want to use the response_curves function instead. It basically is identical to the tuning_curves function, except that it assumes that the input is already encoded, so the encoder is ignored in the plots.
As a side note on terminology, we use the term “tuning curve” to describe the range of the neuron’s activity given a signal in vector space. This means that you can have multi-dimensional tuning curves (since you can feed in multi-dimensional vectors into ensembles). “Response curves”, on the other hand, is used to describe the mapping between some input “current” (i.e., vector input x encoders) and the neuron activity.
@xchoo so in a way the Response Curve is basically a Tuning Curve with all encoders set to 1 ?
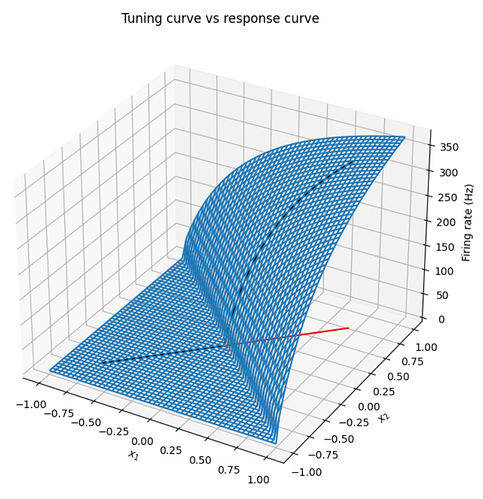
Nooooooot quite, but almost. You can think of the neuron’s response curve as a 1D slice through a neuron’s multi-dimensional tuning curve. And this slice is done in the direction of the neuron’s encoding vector. Below is an example of a 2D ensemble with 1 neuron.
The plot below shows the tuning curve vs response curve of a single neuron with an intercept at 0, and an encoder of [0.707], [0.707]. The encoder is shown as the red line on the xy plane, the neuron’s tuning curve is the blue surface, and the neuron’s response curve is the black line. Notice how the response curve is simply the slice of the neuron’s tuning curve along the neuron’s encoder. I’ve attached the code below if you are interested in playing with it.
test_tuningcurves_2D.py (1.3 KB)
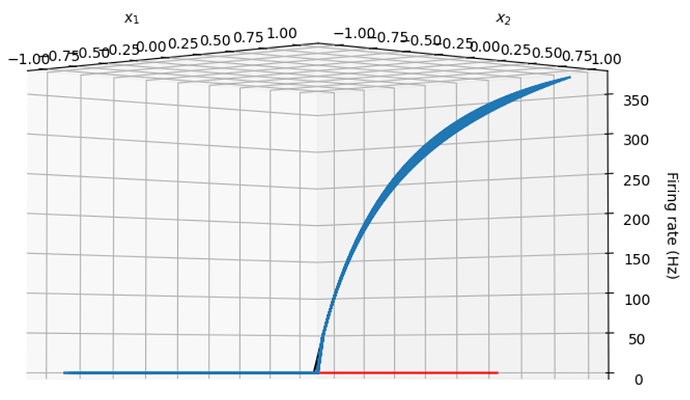
If you adjust the plot so that you are looking directly perpendicular to the neuron’s encoders, the relationship between tuning curve and response curve becomes more clear
Note that the tuning curve is plotted in a -1 to 1 grid, whereas the encoder is normalized to unit length, which is why it looks a little shorter.
In the single dimensional case, the encoding vectors are either [1] or [-1]. Travelling in the direction of an encoder that is [-1] is simply running along the x-axis in the reverse direction, so to generate the response curves (for a 1D ensemble), it’s equivalent to just flipping the tuning curves of any neuron with an encoder of [-1] about the x-axis, which gives the impression that all of the encoders are 1.
Did this get fixed or the error message improved following Fix validation of Choice dist with scalar samples by drasmuss · Pull Request #1630 · nengo/nengo · GitHub?
No. The error message I pasted above is using the latest Nengo master, which according to github already has that PR merged in?
Hey @xchoo, thanks a lot for the help and the valuable information!