Dear Xuan
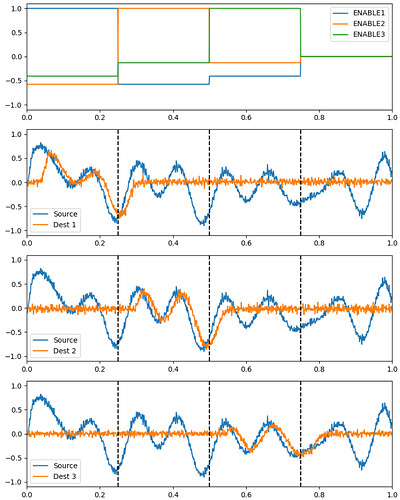
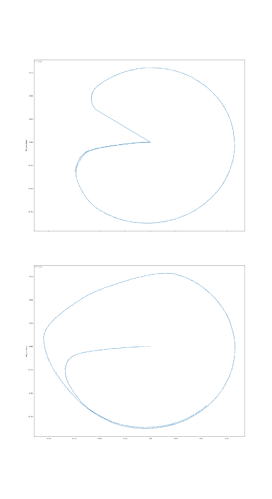
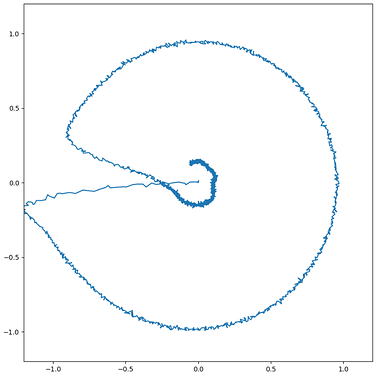
I was able to implement your advised solution 1 (convert ensembles in SPA module. ensembles) and have attached two figures which show a syllable oscillator and the coupled speech actions (just look at row “oscillators” and “speech actions”).
In case of the syllable oscillator for /pi/ (two lines per oscillator → so: only one oscillator is active!) , three actions are activated (via BG-Thal): the vowel action for /i/ → sa_vow_i (magenta), the consonant action for /p/ → sa_constr_lab (labial cnstriction: dark blue) and a action which signals the end of the syllable → sa_final (light blue).
The rest is just concerning the connection of the syllable oscillator with the speech action oscillator for the vowel I (magenta line in row" speech actions")
Now I address the problem of activating the speech actions (here the vowel action):
To my knowledge a direct formulation of the direct neural connection is not possible in the BG-Thal (in the action selection part). The connection is:
nengo.Connection(BI_oscillator.output, sa_vow_I.input, function=CV_signal_sa_vow_I, transform=[vow_vel])
with:
def CV_signal_sa_vow_I(x):
theta = math.atan2(x[1], x[0])
t = theta/ (2*np.pi) + 0.5
return CV_func_sa_vow_I(t)
and:
CV_func_sa_vow_I = piecewise({0:[0], 0.3:[0.7], 0.7:[0]})
Because this connection is thus complex, to my knowledge it can not be written as
BI_oscillator.output >> sa_vow_I.input
and thus can not be used directly in the action selection part of the source code.
Thus, I define two further SPA module ensembles called:
connIn_syll2sa_vow_I = EnsembleModule(ens_params={"n_neurons": n_ens_syll2sa, "dimensions": 2})
connOut_syll2sa_vow_I = EnsembleModule(ens_params={"n_neurons": n_ens_syll2sa, "dimensions": 2})
and I define the connection between syllable oscillator for bi and speech action vor vowel i as:
nengo.Connection(BI_oscillator.output, connIn_syll2sa_vow_I.input)
connIn_syll2sa_vow_I >> connOut_syll2sa_vow_I
nengo.Connection(connIn_syll2sa_vow_I.output, sa_vow_I.input, function=CV_signal_sa_vow_I, transform=[vow_vel])
and now the line
connIn_syll2sa_vow_I >> connOut_syll2sa_vow_I
can be part of the BG-Thal action selection part in the source code.
But doing so (even If I use the source code as written above without running the neural connection
connIn_syll2sa_vow_I >> connOut_syll2sa_vow_I
within the BG-Tal action selection part of the module (but let it run permanently like written above),
I get an artifact, which is:
There is activity of the controlled neuron ensemble (i.e. of the speech action ensemble for vowel i → sa_vow_I) already before the syllable oscillator for bi (-> BI_oscillator) is activated. This activation takes place at around 0.6 sec in the simulation, but we can see activation for the vowel bi speech action (magenta) already earlier between 0 sec and 0.6 sec
This artifact does not appear, if I do not include the ensemble modules
connIn_syll2sa_vow_I = EnsembleModule(ens_params={"n_neurons": n_ens_syll2sa, "dimensions": 2})
connOut_syll2sa_vow_I = EnsembleModule(ens_params={"n_neurons": n_ens_syll2sa, "dimensions": 2})
and thus, if I do the direct connection of syllable oscillator for bi and speech action ensemble for vowel i :
nengo.Connection(BI_oscillator.output, sa_vow_I.input, function=CV_signal_sa_vow_I, transform=[vow_vel])
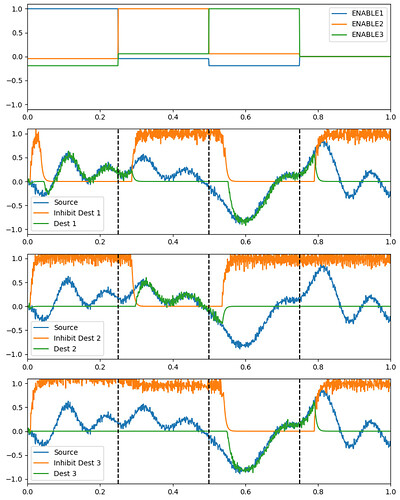
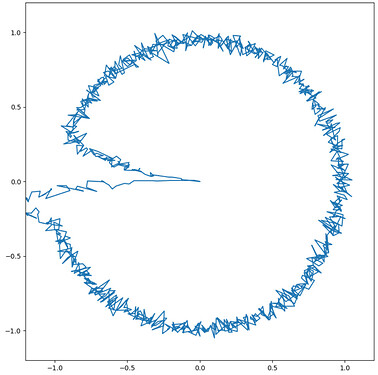
In order to prove that, I as well have attached a second figure which shows the simulation result in case of the direct connection. In this case you see no artifact (no activation of speech action for vowel i before the syllable oscillator becomes active (i.e. see row above in the figure → oscillators).
Do you have any ideas how to solve this problem?
Best
Bernd
In order to give some more details, here the definition for the syllable oscillator (as ensemble module) and for the speech action ensemble module:
# define oscillators for syllables (syll)
n_ens_syll = 2500 # 2500
# Oscillators for each syllable (set intercepts to create a representational deadzone)
BA_oscillator = EnsembleModule(ens_params={"n_neurons": n_ens_syll, "dimensions": 2, "intercepts": Uniform(0.3,1), "encoders": encoders})
DA_oscillator = EnsembleModule(ens_params={"n_neurons": n_ens_syll, "dimensions": 2, "intercepts": Uniform(0.3,1), "encoders": encoders})
GA_oscillator = EnsembleModule(ens_params={"n_neurons": n_ens_syll, "dimensions": 2, "intercepts": Uniform(0.3,1), "encoders": encoders})
PA_oscillator = EnsembleModule(ens_params={"n_neurons": n_ens_syll, "dimensions": 2, "intercepts": Uniform(0.3,1), "encoders": encoders})
TA_oscillator = EnsembleModule(ens_params={"n_neurons": n_ens_syll, "dimensions": 2, "intercepts": Uniform(0.3,1), "encoders": encoders})
KA_oscillator = EnsembleModule(ens_params={"n_neurons": n_ens_syll, "dimensions": 2, "intercepts": Uniform(0.3,1), "encoders": encoders})
MA_oscillator = EnsembleModule(ens_params={"n_neurons": n_ens_syll, "dimensions": 2, "intercepts": Uniform(0.3,1), "encoders": encoders})
NA_oscillator = EnsembleModule(ens_params={"n_neurons": n_ens_syll, "dimensions": 2, "intercepts": Uniform(0.3,1), "encoders": encoders})
LA_oscillator = EnsembleModule(ens_params={"n_neurons": n_ens_syll, "dimensions": 2, "intercepts": Uniform(0.3,1), "encoders": encoders})
# to learn:
BI_oscillator = EnsembleModule(ens_params={"n_neurons": n_ens_syll, "dimensions": 2, "intercepts": Uniform(0.3,1), "encoders": encoders})
DI_oscillator = EnsembleModule(ens_params={"n_neurons": n_ens_syll, "dimensions": 2, "intercepts": Uniform(0.3,1), "encoders": encoders})
GI_oscillator = EnsembleModule(ens_params={"n_neurons": n_ens_syll, "dimensions": 2, "intercepts": Uniform(0.3,1), "encoders": encoders})
PI_oscillator = EnsembleModule(ens_params={"n_neurons": n_ens_syll, "dimensions": 2, "intercepts": Uniform(0.3,1), "encoders": encoders})
TI_oscillator = EnsembleModule(ens_params={"n_neurons": n_ens_syll, "dimensions": 2, "intercepts": Uniform(0.3,1), "encoders": encoders})
KI_oscillator = EnsembleModule(ens_params={"n_neurons": n_ens_syll, "dimensions": 2, "intercepts": Uniform(0.3,1), "encoders": encoders})
MI_oscillator = EnsembleModule(ens_params={"n_neurons": n_ens_syll, "dimensions": 2, "intercepts": Uniform(0.3,1), "encoders": encoders})
NI_oscillator = EnsembleModule(ens_params={"n_neurons": n_ens_syll, "dimensions": 2, "intercepts": Uniform(0.3,1), "encoders": encoders})
LI_oscillator = EnsembleModule(ens_params={"n_neurons": n_ens_syll, "dimensions": 2, "intercepts": Uniform(0.3,1), "encoders": encoders})
and:
n_ens_sa = 2000 # 2000
# Premotor trajectory generators for speech actions "sa"
sa_lab_constr = EnsembleModule(ens_params={"n_neurons": n_ens_sa, "dimensions": 1}) # b, p, m,
sa_api_constr = EnsembleModule(ens_params={"n_neurons": n_ens_sa, "dimensions": 1}) # d, t, n,
sa_dor_constr = EnsembleModule(ens_params={"n_neurons": n_ens_sa, "dimensions": 1}) # g, k, N,
sa_lat_constr = EnsembleModule(ens_params={"n_neurons": n_ens_sa, "dimensions": 1}) # lateral
sa_vel_abduc = EnsembleModule(ens_params={"n_neurons": n_ens_sa, "dimensions": 1}) # nasal
sa_glott_abduc = EnsembleModule(ens_params={"n_neurons": n_ens_sa, "dimensions": 1}) # voiceless sounds
sa_vow_I = EnsembleModule(ens_params={"n_neurons": n_ens_sa, "dimensions": 1}) # vowel I ii
sa_vow_A = EnsembleModule(ens_params={"n_neurons": n_ens_sa, "dimensions": 1}) # vowel A aa
sa_vow_U = EnsembleModule(ens_params={"n_neurons": n_ens_sa, "dimensions": 1}) # vowel U uu
sa_final = EnsembleModule(ens_params={"n_neurons": n_ens_sa, "dimensions": 1}) # end of syllable -> feedback signal
Kind regards
Bernd
www.speechtrainer.eu