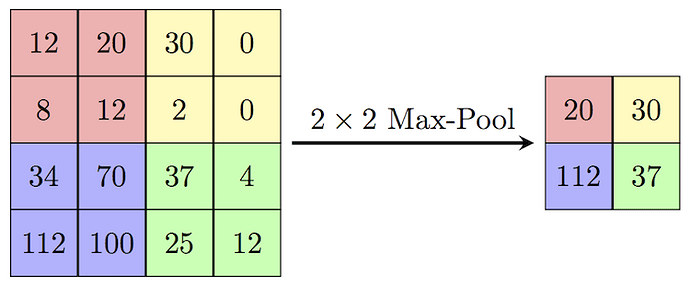Thank you @xchoo for looking into it. I have experimented with your suggested method of replacing a TensorNode and was seemingly successful. However, I have further questions and for the same, I am taking an example of simple 2D CNN network (taken from Nengo-DL examples) and added a MaxPooling op to result in creation of a TensorNode after conversion. Below is the TF network.
# input
inp = tf.keras.Input(shape=(28, 28, 1))
# convolutional layers
conv0 = tf.keras.layers.Conv2D(
filters=32,
kernel_size=3,
activation=tf.nn.relu,
)(inp)
max_pool = tf.keras.layers.MaxPool2D()(conv0)
conv1 = tf.keras.layers.Conv2D(
filters=64,
kernel_size=3,
strides=2,
activation=tf.nn.relu,
)(max_pool)
# fully connected layer
flatten = tf.keras.layers.Flatten()(conv1)
dense = tf.keras.layers.Dense(units=10, activation="softmax")(flatten)
model = tf.keras.Model(inputs=inp, outputs=dense)
Its model.summary() output is below.
Model: "model"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input_1 (InputLayer) [(None, 28, 28, 1)] 0
_________________________________________________________________
conv2d (Conv2D) (None, 26, 26, 32) 320
_________________________________________________________________
max_pooling2d (MaxPooling2D) (None, 13, 13, 32) 0
_________________________________________________________________
conv2d_1 (Conv2D) (None, 6, 6, 64) 18496
_________________________________________________________________
flatten (Flatten) (None, 2304) 0
_________________________________________________________________
dense (Dense) (None, 10) 23050
=================================================================
Total params: 41,866
Trainable params: 41,866
Non-trainable params: 0
_________________________________________________________________
As can be seen above, output from the first conv2d layer is of shape (26, 26, 32) which is input to the max_pooling2d layer. The max-pooling op has a kernel of shape (2, 2) and strides by default is the same, thus the output from the max_pooling2d layer is of shape (13, 13, 32) i.e. image dimensions have been halved and number of filters/channels remain the same. The input to the next conv2d_1 layer (it has a stride of 2) is of the same shape i.e. (13, 13, 32) and thus the output is of shape (6, 6, 64).
Now, let’s try replacing this max_pooling2d TensorNode (after training and conversion). The expectation from the new custom Node would be to accept an input of shape (26, 26, 32) and output data of shape (13, 13, 32) which will be then an input to the next conv2d_1 layer. The operation in the custom Node should respect the topography of the input to it. But how to ensure that Node receives and preserves the correct input topography? Or in other words, how to do the operations in the custom Node which preserves the input/output topography? For the MaxPooling op shown below (source: wikipedia)
I not only have to preserve the above spatial input, but also the filter order (shown below)
How do I preserve the correct input and output topology to/from the custom Node?
For now, I am replacing the max_pooling2d TensorNode with the following code.
def func(t, x):
print(x.shape)
return x[:5408]
with ndl_model.net:
# Create Custom Node.
new_node = nengo.Node(output=func, size_in=21632, label="Custome Node")
conn_from_conv0_to_max_node = ndl_model.net.all_connections[3]
# COnnection from Conv0 to MaxPool node.
nengo.Connection(
conn_from_conv0_to_max_node.pre_obj,
new_node,
transform=conn_from_conv0_to_max_node.transform,
synapse=conn_from_conv0_to_max_node.synapse,
function=conn_from_conv0_to_max_node.function)
# Connection from MaxPool node to Conv1.
conn_from_max_node_to_conv1 = ndl_model.net.all_connections[6]
nengo.Connection(
new_node,
conn_from_max_node_to_conv1.post_obj,
transform=conn_from_max_node_to_conv1.transform,
synapse=conn_from_max_node_to_conv1.synapse,
function=conn_from_max_node_to_conv1.function)
# Remove the old connection to MaxPool node and from MaxPool node, MaxPool node.
ndl_model.net._connections.remove(conn_from_conv0_to_max_node)
ndl_model.net._connections.remove(conn_from_max_node_to_conv1)
ndl_model.net._nodes.remove(conn_from_conv0_to_max_node.post_obj)
I also checked if making the new connection is successful.
ndl_model.net.all_connections
[<Connection at 0x2b97fffc01d0 from <Node "conv2d.0.bias"> to <Node "conv2d.0.bias_relay">>,
<Connection at 0x2b97efeee190 from <Node "conv2d.0.bias_relay"> to <Neurons of <Ensemble "conv2d.0">>>,
<Connection at 0x2b97fffbd990 from <Node "input_1"> to <Neurons of <Ensemble "conv2d.0">>>,
<Connection at 0x2b97fff8b810 from <Node "conv2d_1.0.bias"> to <Node "conv2d_1.0.bias_relay">>,
<Connection at 0x2b97efe79590 from <Node "conv2d_1.0.bias_relay"> to <Neurons of <Ensemble "conv2d_1.0">>>,
<Connection at 0x2b97effc8690 from <Node "dense.0.bias"> to <TensorNode "dense.0">>,
<Connection at 0x2b984b73fad0 from <Neurons of <Ensemble "conv2d_1.0">> to <TensorNode "dense.0">>,
<Connection at 0x2b9b8a11b490 from <Neurons of <Ensemble "conv2d.0">> to <Node "Custome Node">>,
<Connection at 0x2b9b8a12ab90 from <Node "Custome Node"> to <Neurons of <Ensemble "conv2d_1.0">>>]
and from the above output, it seems it is. Also, the output of print(x.shape) in the func() passed in the custom Node is (21632,) = 26 x 26 x 32 is coalesced in one vector. Interestingly, why is the order of all the connections not sequential?
I also probed the ndl_model.net first Conv layer with following code:
with ndl_model.net:
# Output from the first Conv layer.
# ndl_model.layers[conv0].probeable => ('output', 'input', 'output', 'voltage')
conv0_lyr_otpt = nengo.Probe(ndl_model.layers[conv0], attr="output")
# ndl_model.net.ensembles[0].probeable => ('decoded_output', 'input', 'scaled_encoders')
conv0_ens_otpt = nengo.Probe(ndl_model.net.ensembles[0], attr="decoded_output")
# ndl_model.layers[conv0].ensemble.neurons.probeable => ('output', 'input', 'output', 'voltage')
conv0_lyr_nrns_otpt = nengo.Probe(ndl_model.layers[conv0].ensemble.neurons, attr="output")
# ndl_model.net.ensembles[0].neurons.probeable => ('output', 'input', 'output', 'voltage')
conv0_ens_nrns_otpt = nengo.Probe(ndl_model.net.ensembles[0].neurons, attr="output")
I was just curious about the output from ndl_model.layers and ndl_model.net.ensembles from the first conv2d layer and it seems that the outputs from both sources match, as shown below… so they are essentially representing the same thing… right?
neuron_index = 16345
print(data1[conv0_lyr_otpt][0, :, neuron_index])
print(data1[conv0_lyr_nrns_otpt][0, :, neuron_index])
print(data1[conv0_ens_nrns_otpt][0, :, neuron_index])
[0. 0. 0. 9.999999 0. 0. 0. 0.
9.999999 0. 0. 0. 9.999999 0. 0. 0.
0. 9.999999 0. 0. 0. 9.999999 0. 0.
0. 0. 9.999999 0. 0. 0. 0. 9.999999
0. 0. 0. 9.999999 0. 0. 0. 0. ]
[0. 0. 0. 9.999999 0. 0. 0. 0.
9.999999 0. 0. 0. 9.999999 0. 0. 0.
0. 9.999999 0. 0. 0. 9.999999 0. 0.
0. 0. 9.999999 0. 0. 0. 0. 9.999999
0. 0. 0. 9.999999 0. 0. 0. 0. ]
[0. 0. 0. 9.999999 0. 0. 0. 0.
9.999999 0. 0. 0. 9.999999 0. 0. 0.
0. 9.999999 0. 0. 0. 9.999999 0. 0.
0. 0. 9.999999 0. 0. 0. 0. 9.999999
0. 0. 0. 9.999999 0. 0. 0. 0. ]
except that conv0_ens_otpt = nengo.Probe(ndl_model.net.ensembles[0], attr="decoded_output") produces a output of shape (n_test_images, n_steps, 1), i.e. perhaps the overall decoded output from the Ensemble of 21632 neurons. Right?
One interesting thing I noted from above output was that the inputs to the max_pooling2d layer are the scaled spikes (I thought the inputs should have been smoothed values), but I guess, it goes with what you mentioned earlier, the outputs from the Ensemble neurons are the spikes and the smoothing happens during the input to the next Ensemble, and since MaxPooling2D op is not an Ensemble it simply operates on scaled spikes every timestep e.g. max(9.9999, 0, 0, 0) = 9.9999 i.e. simply output the max amplitude spike as an input to the next Ensemble… right?
Sorry for such a long response, please let me know if anything is confusing here.