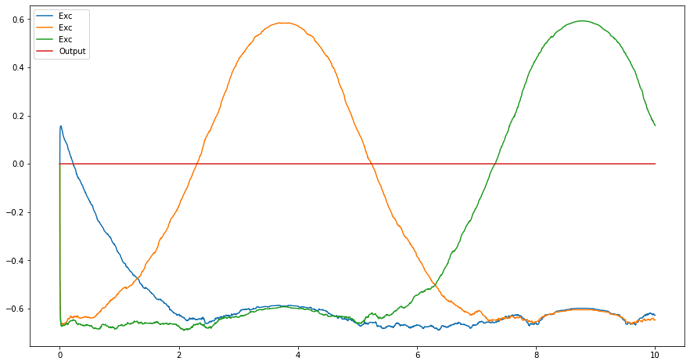
Hello,
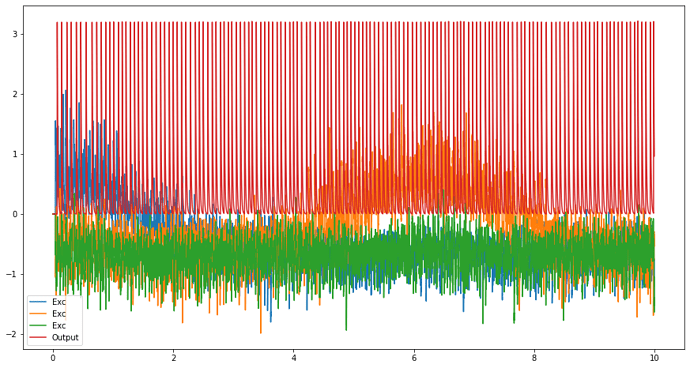
I’d like to use a set of weights from a decoded connection in a direct connection; the tuning curves for the ensemble look correct, however a simulation with a direct connection and the decoded weights does not produce correct output. Below is my code, and I would be very grateful if someone could let me know what I am doing wrong. Thanks!
import numpy as np
import matplotlib.pyplot as plt
from matplotlib import cm
import nengo
from nengo.utils.least_squares_solvers import SVD
from scipy.interpolate import pchip
def gaussian(x, mu, sig):
return np.exp(-np.power(x - mu, 2.) / (2 * np.power(sig, 2.)))
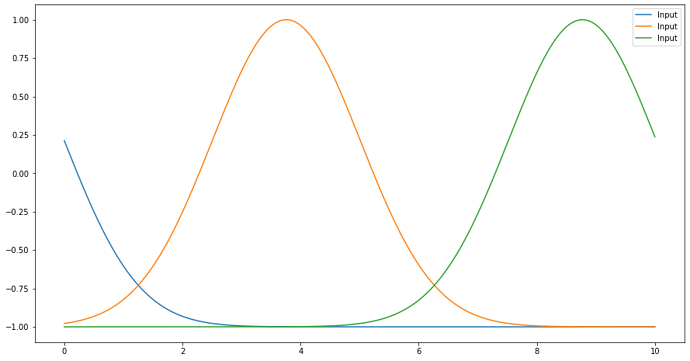
# Inputs mimicking receptive fields in a linear space
N = 1000
width = N/20 # receptive field width
input_rates = []
input_space = np.arange(N)
for i in range(3):
loc = float(i+1)*N/4 # peak of the receptive field of each input
input_rate = gaussian(input_space, loc, width)
input_rates.append(input_rate)
input_matrix = np.column_stack(input_rates)
target_loc = N/2 # peak of the receptive field we wish to encode
target_rate = gaussian(input_space, target_loc, width)
trajectory_x = np.arange(250, 650)
trajectory_t = np.linspace(0., 10., len(trajectory_x))
trajectory_inputs = []
for input_rate in input_rates:
trajectory_rate = np.interp(trajectory_x, input_space, input_rate)
trajectory_input = pchip(trajectory_t, trajectory_rate)
trajectory_inputs.append(trajectory_input)
# Target function
target_trajectory_rate = np.interp(trajectory_x, input_space, target_rate)
target_trajectory_input = pchip(trajectory_t, target_trajectory_rate)
# Representation of inputs on a trajectory through the input space
def trajectory_function(t):
result = np.asarray([ 2.*y(t) - 1. for y in trajectory_inputs ])
return result
X_train = 2.*input_matrix - 1.
y_train = target_rate.reshape((-1,1))
X_test = trajectory_function(trajectory_t).T
y_test = target_trajectory_input(trajectory_t).ravel()
n_features = X_train.shape[1]
n_output = y_train.shape[1]
solver = nengo.solvers.LstsqL2nz(reg=0.01, solver=SVD())
ens_params = dict(
neuron_type=nengo.LIF(),
intercepts=nengo.dists.Choice([0.0]),
max_rates=nengo.dists.Choice([20])
)
# obtain decoder weights for the given inputs and target receptive field
with nengo.Network(seed=19, label="Receptive field tuning") as tuning_model:
tuning_model.PC = nengo.Ensemble(n_neurons=50, dimensions=n_features,
eval_points=X_train,
**ens_params)
tuning_model.output = nengo.Ensemble(n_neurons=1, dimensions=1,
**ens_params)
PC_to_output = nengo.Connection(tuning_model.PC,
tuning_model.output.neurons,
synapse=None,
eval_points=X_train,
function=y_train,
solver=solver)
tuning_sim = nengo.Simulator(tuning_model)
output_weights = tuning_sim.data[PC_to_output].weights
_, train_acts = nengo.utils.ensemble.tuning_curves(tuning_model.PC, tuning_sim, inputs=X_train)
_, test_acts = nengo.utils.ensemble.tuning_curves(tuning_model.PC, tuning_sim, inputs=X_test)
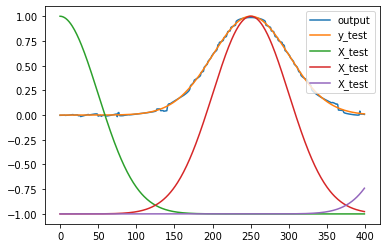
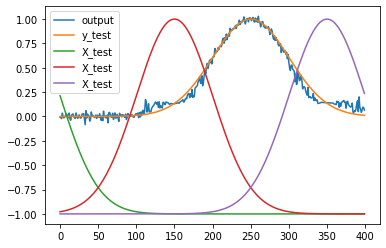
plt.figure()
plt.plot(y_train)
plt.plot(np.dot(train_acts, output_weights.T))
plt.figure()
plt.plot(y_test)
plt.plot(np.dot(test_acts, output_weights.T))
# use the decoded weights in a direct connection
network_weights = tuning_sim.data[PC_to_output].weights
with nengo.Network(label="Basic network model", seed=19) as model:
model.Input = nengo.Node(trajectory_function, size_out=X_train.shape[1])
model.Exc = nengo.Ensemble(50, dimensions=X_train.shape[1],
**ens_params)
model.Output = nengo.Ensemble(1, dimensions=1,
**ens_params)
nengo.Connection(model.Input, model.Exc)
nengo.Connection(model.Exc.neurons, model.Output,
transform=network_weights,
synapse=0.01)
with model:
Input_probe = nengo.Probe(model.Input, synapse=0.01)
Exc_probe = nengo.Probe(model.Exc, synapse=0.01)
Output_probe = nengo.Probe(model.Output, synapse=0.01)
with nengo.Simulator(model) as sim:
sim.run(np.max(trajectory_t))
plt.figure(figsize=(15,8))
#plt.plot(sim.trange(), sim.data[Input_probe], label='Input')
plt.plot(sim.trange(), sim.data[Exc_probe], label='Exc')
plt.plot(sim.trange(), sim.data[Output_probe], label='Output')
plt.legend();