Hello Aaron! Sure thing.
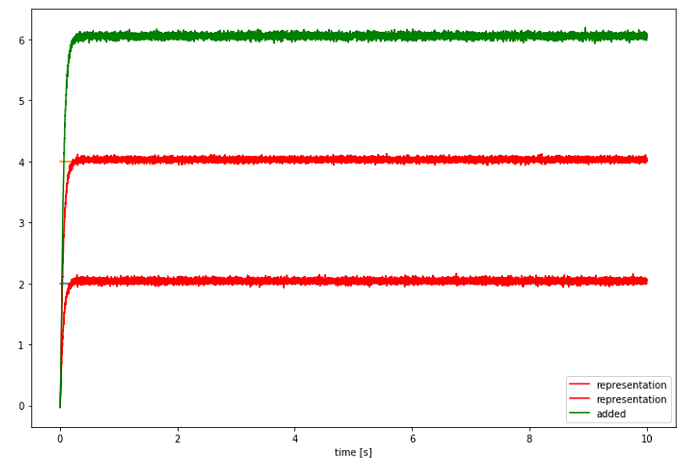
Here, I’m adopting the addition example from the Nengo Core documentation. Here is the network and output when using the core simulator in Jupyter:
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import nengo
import nengo_loihi
with nengo.Network(label="Addition") as model:
inp1 = nengo.Node(output=2)
inp2 = nengo.Node(output=4)
A = nengo.Ensemble(200, dimensions=2, label="Input Ensemble", radius=10)
B = nengo.Ensemble(100, dimensions=1, label="Output Ensemble", radius=10)
nengo.Connection(inp1, A[0])
nengo.Connection(inp2, A[1])
nengo.Connection(A, B, function=lambda x: x[0] + x[1])
inp1_p = nengo.Probe(inp1, 'output', label="input 1")
inp2_p = nengo.Probe(inp2, 'output', label="input 2")
A_p = nengo.Probe(A, 'decoded_output', synapse=0.05, label="representation")
B_p = nengo.Probe(B, 'decoded_output', synapse=0.05, label="added")
with nengo.Simulator(model) as sim:
sim.run(10)
# Plot the decoded output of the ensemble
plt.figure(figsize=(12,8))
plt.plot(sim.trange(), sim.data[inp1_p], label=inp1.label)
plt.plot(sim.trange(), sim.data[inp2_p], label=inp2.label)
plt.plot(sim.trange(), sim.data[A_p], 'r', label=A_p.label)
plt.plot(sim.trange(), sim.data[B_p], 'g', label=B_p.label)
plt.legend()
plt.xlabel('time [s]');
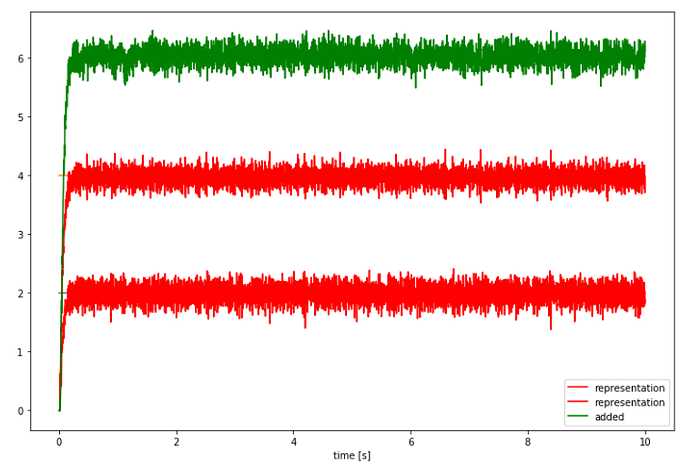
Now, when trying to run on Loihi:
nengo_loihi.set_defaults()
with nengo.Network(label="Addition") as model:
inp1 = nengo.Node(output=2)
inp2 = nengo.Node(output=4)
A = nengo.Ensemble(200, dimensions=2, label="Input Ensemble", radius=10)
B = nengo.Ensemble(100, dimensions=1, label="Output Ensemble", radius=10)
nengo.Connection(inp1, A[0])
nengo.Connection(inp2, A[1])
nengo.Connection(A, B, function=lambda x: x[0] + x[1])
inp1_p = nengo.Probe(inp1, 'output', label="input 1")
inp2_p = nengo.Probe(inp2, 'output', label="input 2")
A_p = nengo.Probe(A, 'decoded_output', synapse=0.05, label="representation")
B_p = nengo.Probe(B, 'decoded_output', synapse=0.05, label="added")
with nengo_loihi.Simulator(model) as sim:
sim.run(10)
Does the end of this stack trace indicate that I have to include encoders for each of the ensembles? If so, how?
Then, if I try and represent in each of the inputs in their own dedicated ensembles instead of trying to represent both in A:
nengo_loihi.set_defaults()
with nengo.Network(label="Addition") as model:
inp1 = nengo.Node(output=2)
inp2 = nengo.Node(output=4)
#A = nengo.Ensemble(200, dimensions=2, label="Input Ensemble", radius=10)
A = nengo.networks.EnsembleArray( 200,
n_ensembles=2,
ens_dimensions=1,
radius= 10 )
B = nengo.Ensemble(100, dimensions=1, label="Output Ensemble", radius=10)
A.output.output = lambda t, x: x
nengo.Connection(inp1, A.ea_ensembles[0])
nengo.Connection(inp2, A.ea_ensembles[1])
nengo.Connection(A.output, B, function=lambda x: x[0] + x[1])
inp1_p = nengo.Probe(inp1, 'output', label="input 1")
inp2_p = nengo.Probe(inp2, 'output', label="input 2")
A_p = nengo.Probe(A.output, synapse=0.05, label="representation")
B_p = nengo.Probe(B, 'decoded_output', synapse=0.05, label="added")
with nengo_loihi.Simulator(model) as sim:
sim.run(10)
…
This leads me to think that when using Loihi as the backend, one must dedicate entire ensembles to each value that one would like to use. Is this true?
Also, when applying a function on a connection, how can I incorporate other inputs (aside from using transform to apply weights)? For example, say I wanted to apply this function across a connection from x to some other ensemble:
def my_func(x, value):
return x + 2*value
How could I include the second argument? So far, I’ve only been able to get a connection function to take in a single argument, being the input object of the connection. Two scenarios where I might want to include value is when
-
value is the decoded output of a separate ensemble
-
value is the output from a node
An immediate remedy seems to try breaking down the computation across multiple objects, but I thought you might have a recommendation to more easily implement this.
Hopefully that clarifies some things! I will same remaining questions for later in the thread since this post is so long. 
Thanks!